Robust inference of the context specific structure and temporal dynamics of gene regulatory network
- PMID: 21143778
- PMCID: PMC2999341
- DOI: 10.1186/1471-2164-11-S3-S11
Robust inference of the context specific structure and temporal dynamics of gene regulatory network
Abstract
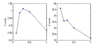
Background: Response of cells to changing endogenous or exogenous conditions is governed by intricate molecular interactions, or regulatory networks. To lead to appropriate responses, regulatory network should be 1) context-specific, i.e., its constituents and topology depend on the phonotypical and experimental context including tissue types and cell conditions, such as damage, stress, macroenvironments of cell, etc. and 2) time varying, i.e., network elements and their regulatory roles change actively over time to control the endogenous cell states e.g. different stages in a cell cycle.
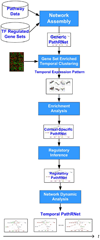
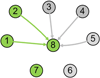
Results: A novel network model PathRNet and a reconstruction approach PATTERN are proposed for reconstructing the context specific time varying regulatory networks by integrating microarray gene expression profiles and existing knowledge of pathways and transcription factors. The nodes of the PathRNet are Transcription Factors (TFs) and pathways, and edges represent the regulation between pathways and TFs. The reconstructed PathRNet for Kaposi's sarcoma-associated herpesvirus infection of human endothelial cells reveals the complicated dynamics of the underlying regulatory mechanisms that govern this intricate process. All the related materials including source code are available at http://compgenomics.utsa.edu/tvnet.html.
Conclusions: The proposed PathRNet provides a system level landscape of the dynamics of gene regulatory circuitry. The inference approach PATTERN enables robust reconstruction of the temporal dynamics of pathway-centric regulatory networks. The proposed approach for the first time provides a dynamic perspective of pathway, TF regulations, and their interaction related to specific endogenous and exogenous conditions.
Figures
















References
-
- Van Someren EP, Wessels LF, Backer E, Reinders MJ. "Genetic network modeling,". Pharmacogenomics. 2002;vol. 3:pp. 507–25. - PubMed
-
- Shmulevich I, Dougherty ER, Kim S, Zhang W. "Probabilistic Boolean Networks: a rule-based uncertainty model for gene regulatory networks,". Bioinformatics. 2002;vol. 18:pp. 261–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

