Probing the anticancer mechanism of prospective herbal drug Withaferin A on mammals: a case study on human and bovine proteasomes
- PMID: 21143798
- PMCID: PMC3005937
- DOI: 10.1186/1471-2164-11-S4-S15
Probing the anticancer mechanism of prospective herbal drug Withaferin A on mammals: a case study on human and bovine proteasomes
Abstract
Background: The UPP (ubiquitin proteasome pathway) is the major proteolytic system in the cytosol and nucleus of all eukaryotic cells which regulates cellular events, including mitotis, differentiation, signal transduction, apoptosis, and inflammation. UPP controls activation of the transcriptional factor NF-κB (nuclear factor κB), which is a regulatory protein playing central role in a variety of cellular processes including immune and inflammatory responses, apoptosis, and cellular proliferation. Since the primary interaction of proteasomes occurs with endogenous proteins, the signalling action of transcription factor NF-κB can be blocked by inhibition of proteasomes. A great variety of natural and synthetic chemical compounds classified as peptide aldehydes, peptide boronates, nonpeptide inhibitors, peptide vinyl sulfones and epoxyketones are now widely used as research tools for probing their potential to inhibit proteolytic activities of different proteasomes and to investigate the underlying inhibition mechanisms. The present work reports a bio-computational study carried out with the aim of exploring the proteasome inhibition capability of WA (withaferin A), a steroidal lactone, by understanding the binding mode of WA as a ligand into the mammalian proteasomes (X-ray crystal structure of Bos taurus 20S proteasome and multiple template homology modelled structure of 20S proteasome of Homo sapiens) using molecular docking and molecular dynamics simulation studies.
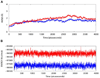
Results: One possible mode of action which is proposed here for WA to act as a proteasome inhibitor is by suppression of the proteolytic activity which depends on the N-terminal threonine (Thr1) residue hydroxyl group. Docking studies carried out with herbal ligand WA into the structures of bovine and human proteasomes substantiate that WA has the ability to inhibit activity of mammalian 20S proteasomes by blocking the nucleophilic function of N-terminal Thr1. Results from molecular dynamics simulations in water show that the trajectories of both the native human 20S proteasome and the proteasome complexed with WA are stable over a considerably long time period of 4 ns suggesting the dynamic structural stability of human 20S proteasome/WA complex.
Conclusions: Inhibition of proteasomal activity are promising ways to retard or block degradation of specific proteins to correct diverse pathologies. Though quite a number of selective and efficient proteasomal inhibitors exist nowadays, their toxic side effects limit their potential in possible disease treatment. Thus there is an indispensable need for exploration of novel natural products as antitumor drug candidates. The present work supports the mammalian proteasomes inhibiting activity of WA along with elucidation of its possible mode of action. Since WA is a small herbal molecule, it is expected to provide one of the modest modes of inhibition along with added favours of ease in oral administration and decreased immunogenicity. The molecular docking results suggest that WA can inhibit the mammalian proteasomes irreversibly and with a high rate through acylation of the N-terminal Thr1 of the β-5 subunit.
Figures






Similar articles
-
Inhibition of the NEMO/IKKβ association complex formation, a novel mechanism associated with the NF-κB activation suppression by Withania somnifera's key metabolite withaferin A.BMC Genomics. 2010 Dec 2;11 Suppl 4(Suppl 4):S25. doi: 10.1186/1471-2164-11-S4-S25. BMC Genomics. 2010. PMID: 21143809 Free PMC article.
-
The tumor proteasome is a primary target for the natural anticancer compound Withaferin A isolated from "Indian winter cherry".Mol Pharmacol. 2007 Feb;71(2):426-37. doi: 10.1124/mol.106.030015. Epub 2006 Nov 8. Mol Pharmacol. 2007. PMID: 17093135
-
Hsp90/Cdc37 chaperone/co-chaperone complex, a novel junction anticancer target elucidated by the mode of action of herbal drug Withaferin A.BMC Bioinformatics. 2011 Feb 15;12 Suppl 1(Suppl 1):S30. doi: 10.1186/1471-2105-12-S1-S30. BMC Bioinformatics. 2011. PMID: 21342561 Free PMC article.
-
The ubiquitin-proteasome system as a prospective molecular target for cancer treatment and prevention.Curr Protein Pept Sci. 2010 Sep;11(6):459-70. doi: 10.2174/138920310791824057. Curr Protein Pept Sci. 2010. PMID: 20491623 Free PMC article. Review.
-
[Ubiquitin-independent protein degradation in proteasomes].Biomed Khim. 2018 Mar;64(2):134-148. doi: 10.18097/PBMC20186402134. Biomed Khim. 2018. PMID: 29723144 Review. Russian.
Cited by
-
Withaferin A suppresses skin tumor promotion by inhibiting proteasome-dependent isocitrate dehydrogenase 1 degradation.Transl Cancer Res. 2019 Oct;8(6):2449-2460. doi: 10.21037/tcr.2019.09.57. Transl Cancer Res. 2019. PMID: 35116997 Free PMC article.
-
Effect of Withaferin-A, Withanone, and Caffeic Acid Phenethyl Ester on DNA Methyltransferases: Potential in Epigenetic Cancer Therapy.Curr Top Med Chem. 2024;24(4):379-391. doi: 10.2174/1568026623666230726105017. Curr Top Med Chem. 2024. PMID: 37496252
-
Ashwagandha derived withanone targets TPX2-Aurora A complex: computational and experimental evidence to its anticancer activity.PLoS One. 2012;7(1):e30890. doi: 10.1371/journal.pone.0030890. Epub 2012 Jan 27. PLoS One. 2012. PMID: 22303466 Free PMC article.
-
Blocking protein kinase C signaling pathway: mechanistic insights into the anti-leishmanial activity of prospective herbal drugs from Withania somnifera.BMC Genomics. 2012;13 Suppl 7(Suppl 7):S20. doi: 10.1186/1471-2164-13-S7-S20. Epub 2012 Dec 13. BMC Genomics. 2012. PMID: 23281834 Free PMC article.
-
Differential activities of the two closely related withanolides, Withaferin A and Withanone: bioinformatics and experimental evidences.PLoS One. 2012;7(9):e44419. doi: 10.1371/journal.pone.0044419. Epub 2012 Sep 4. PLoS One. 2012. PMID: 22973447 Free PMC article.
References
-
- Peters JM, Franke WW, Kleinschmidt JA. Distinct 19-S and 20-S Subcomplexes of the 26-S Proteasome and Their Distribution in the Nucleus and the Cytoplasm. J Biol Chem. 1994;269(10):7709–7718. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

