Disease-associated alleles in genome-wide association studies are enriched for derived low frequency alleles relative to HapMap and neutral expectations
- PMID: 21143973
- PMCID: PMC3017004
- DOI: 10.1186/1755-8794-3-57
Disease-associated alleles in genome-wide association studies are enriched for derived low frequency alleles relative to HapMap and neutral expectations
Abstract
Background: Genome-wide association studies give insight into the genetic basis of common diseases. An open question is whether the allele frequency distributions and ancestral vs. derived states of disease-associated alleles differ from the rest of the genome. Characteristics of disease-associated alleles can be used to increase the yield of future studies.
Methods: The set of all common disease-associated alleles found in genome-wide association studies prior to January 2010 was analyzed and compared with HapMap and theoretical null expectations. In addition, allele frequency distributions of different disease classes were assessed. Ages of HapMap and disease-associated alleles were also estimated.
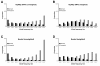
Results: The allele frequency distribution of HapMap alleles was qualitatively similar to neutral expectations. However, disease-associated alleles were more likely to be low frequency derived alleles relative to null expectations. 43.7% of disease-associated alleles were ancestral alleles. The mean frequency of disease-associated alleles was less than randomly chosen CEU HapMap alleles (0.394 vs. 0.610, after accounting for probability of detection). Similar patterns were observed for the subset of disease-associated alleles that have been verified in multiple studies. SNPs implicated in genome-wide association studies were enriched for young SNPs compared to randomly selected HapMap loci. Odds ratios of disease-associated alleles tended to be less than 1.5 and varied by frequency, confirming previous studies.
Conclusions: Alleles associated with genetic disease differ from randomly selected HapMap alleles and neutral expectations. The evolutionary history of alleles (frequency and ancestral vs. derived state) influences whether they are implicated in genome-wide association studies.
Figures

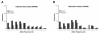


References
-
- Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W. et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nature Genetics. 2007;39(8):984–988. doi: 10.1038/ng2085. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

