BIGSdb: Scalable analysis of bacterial genome variation at the population level
- PMID: 21143983
- PMCID: PMC3004885
- DOI: 10.1186/1471-2105-11-595
BIGSdb: Scalable analysis of bacterial genome variation at the population level
Abstract
Background: The opportunities for bacterial population genomics that are being realised by the application of parallel nucleotide sequencing require novel bioinformatics platforms. These must be capable of the storage, retrieval, and analysis of linked phenotypic and genotypic information in an accessible, scalable and computationally efficient manner.
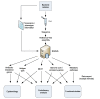
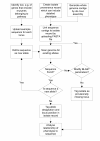
Results: The Bacterial Isolate Genome Sequence Database (BIGSDB) is a scalable, open source, web-accessible database system that meets these needs, enabling phenotype and sequence data, which can range from a single sequence read to whole genome data, to be efficiently linked for a limitless number of bacterial specimens. The system builds on the widely used mlstdbNet software, developed for the storage and distribution of multilocus sequence typing (MLST) data, and incorporates the capacity to define and identify any number of loci and genetic variants at those loci within the stored nucleotide sequences. These loci can be further organised into 'schemes' for isolate characterisation or for evolutionary or functional analyses. Isolates and loci can be indexed by multiple names and any number of alternative schemes can be accommodated, enabling cross-referencing of different studies and approaches. LIMS functionality of the software enables linkage to and organisation of laboratory samples. The data are easily linked to external databases and fine-grained authentication of access permits multiple users to participate in community annotation by setting up or contributing to different schemes within the database. Some of the applications of BIGSDB are illustrated with the genera Neisseria and Streptococcus.The BIGSDB source code and documentation are available at http://pubmlst.org/software/database/bigsdb/.
Conclusions: Genomic data can be used to characterise bacterial isolates in many different ways but it can also be efficiently exploited for evolutionary or functional studies. BIGSDB represents a freely available resource that will assist the broader community in the elucidation of the structure and function of bacteria by means of a population genomics approach.
Figures


References
-
- Baker S, Holt K, van de Vosse E, Roumagnac P, Whitehead S, King E, Ewels P, Keniry A, Weill FX, Lightfoot D. et al. High-throughput genotyping of Salmonella enterica serovar Typhi allowing geographical assignment of haplotypes and pathotypes within an urban District of Jakarta, Indonesia. J Clin Microbiol. 2008;46(5):1741–1746. doi: 10.1128/JCM.02249-07. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

