The folding of single domain proteins--have we reached a consensus?
- PMID: 21144739
- PMCID: PMC3039110
- DOI: 10.1016/j.sbi.2010.11.002
The folding of single domain proteins--have we reached a consensus?
Abstract
Rather than stressing the most recent advances in the field, this review highlights the fundamental topics where disagreement remains and where adequate experimental data are lacking. These topics include properties of the denatured state and the role of residual structure, the nature of the fundamental steps and barriers, the extent of pathway heterogeneity and non-native interactions, recent comparisons between theory and experiment, and finally, dynamical properties of the folding reaction.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
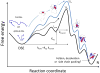
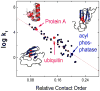

References
-
- McCarney ER, Kohn JE, Plaxco KW. Is there or isn’t there? The case for (and against) residual structure in chemically denatured proteins. Crit Rev Biochem Mol Biol. 2005;40:181–189. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

