Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors
- PMID: 21145483
- PMCID: PMC3023311
- DOI: 10.1016/j.molcel.2010.09.029
Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors
Abstract
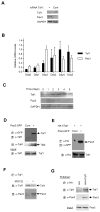
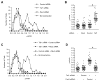
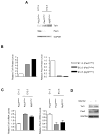
Pax3 plays critical roles during developmental and postnatal myogenesis. We have previously shown that levels of Pax3 protein are regulated by monoubiquitination and proteasomal degradation during postnatal myogenesis, but none of the key regulators of the monoubiquitination process were known. Here we show that Pax3 monoubiquitination is mediated by the ubiquitin-activating/conjugating activity of Taf1, a component of the core transcriptional machinery that was recently reported to be downregulated during myogenic differentiation. We show that Taf1 binds directly to Pax3 and overexpression of Taf1 increases the level of monoubiquitinated Pax3 and its degradation by the proteasome. A decrease of Taf1 results in a decrease in Pax3 monoubiquitination, an increase in the levels of Pax3 protein, and a concomitant increase in Pax3-mediated inhibition of myogenic differentiation and myoblast migration. These results suggest that Taf1 regulates Pax3 protein levels through its ability to mediate monoubiquitination, revealing a critical interaction between two proteins that are involved in distinct aspects of myogenic differentiation. Finally, these results suggest that the components of the core transcriptional are integrally involved in the process of myogenic differentiation, acting as nodal regulators of the differentiation program.
Copyright © 2010 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures




References
-
- Belz T, Pham AD, Beisel C, Anders N, Bogin J, Kwozynski S, Sauer F. In vitro assays to study protein ubiquitination in transcription. Methods. 2002;26:233–244. - PubMed
-
- Biressi S, Molinaro M, Cossu G. Cellular heterogeneity during vertebrate skeletal muscle development. Dev Biol. 2007;308:281–293. - PubMed
-
- Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376:768–771. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

