An ontology-based comparative anatomy information system
- PMID: 21146377
- PMCID: PMC3055271
- DOI: 10.1016/j.artmed.2010.10.001
An ontology-based comparative anatomy information system
Abstract
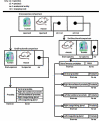
Introduction: This paper describes the design, implementation, and potential use of a comparative anatomy information system (CAIS) for querying on similarities and differences between homologous anatomical structures across species, the knowledge base it operates upon, the method it uses for determining the answers to the queries, and the user interface it employs to present the results. The relevant informatics contributions of our work include (1) the development and application of the structural difference method, a formalism for symbolically representing anatomical similarities and differences across species; (2) the design of the structure of a mapping between the anatomical models of two different species and its application to information about specific structures in humans, mice, and rats; and (3) the design of the internal syntax and semantics of the query language. These contributions provide the foundation for the development of a working system that allows users to submit queries about the similarities and differences between mouse, rat, and human anatomy; delivers result sets that describe those similarities and differences in symbolic terms; and serves as a prototype for the extension of the knowledge base to any number of species. Additionally, we expanded the domain knowledge by identifying medically relevant structural questions for the human, the mouse, and the rat, and made an initial foray into the validation of the application and its content by means of user questionnaires, software testing, and other feedback.
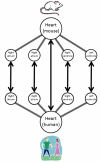
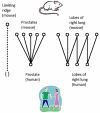
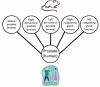
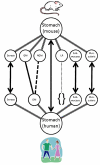
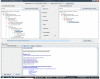
Methods: The anatomical structures of the species to be compared, as well as the mappings between species, are modeled on templates from the Foundational Model of Anatomy knowledge base, and compared using graph-matching techniques. A graphical user interface allows users to issue queries that retrieve information concerning similarities and differences between structures in the species being examined. Queries from diverse information sources, including domain experts, peer-reviewed articles, and reference books, have been used to test the system and to illustrate its potential use in comparative anatomy studies.
Results: 157 test queries were submitted to the CAIS system, and all of them were correctly answered. The interface was evaluated in terms of clarity and ease of use. This testing determined that the application works well, and is fairly intuitive to use, but users want to see more clarification of the meaning of the different types of possible queries. Some of the interface issues will naturally be resolved as we refine our conceptual model to deal with partial and complex homologies in the content.
Conclusions: The CAIS system and its associated methods are expected to be useful to biologists and translational medicine researchers. Possible applications range from supporting theoretical work in clarifying and modeling ontogenetic, physiological, pathological, and evolutionary transformations, to concrete techniques for improving the analysis of genotype-phenotype relationships among various animal models in support of a wide array of clinical and scientific initiatives.
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures




Comment in
-
Translation and localization of SNOMED CT in China: a pilot study.Artif Intell Med. 2012 Feb;54(2):147-9. doi: 10.1016/j.artmed.2011.12.002. Epub 2012 Jan 2. Artif Intell Med. 2012. PMID: 22217426 No abstract available.
References
-
- Biering SF. Evidence-based medicine in treatment and rehabilitation of spinal cord injured. Spinal Cord. 2005 October;43(10):587–92. - PubMed
-
- Eckman BA, Kosky AS, Laroco LA., Jr Extending traditional query-based integration approaches for functional characterization of post-genomic data. Bioinformatics. 2001 July;17(7):587–601. - PubMed
-
- Pagon RA, Tarczy-Hornoch P, Baskin PK, Edwards JE, Covington ML, Espeseth M, et al. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum Mutat. 2002 May;19(5):501–9. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

