The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17
- PMID: 21146513
- PMCID: PMC3461954
- DOI: 10.1016/j.ydbio.2010.12.007
The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17
Abstract
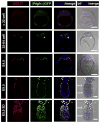
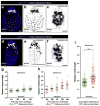
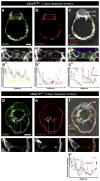
Cells of the primitive endoderm (PrE) and the pluripotent epiblast (EPI), the two lineages specified within the inner cell mass (ICM) of the mouse blastocyst stage embryo, are segregated into adjacent tissue layers by the end of the preimplantation period. The PrE layer which emerges as a polarized epithelium adjacent to the blastocoel, with a basement membrane separating it from the EPI, has two derivatives, the visceral and parietal endoderm. In this study we have investigated the localization of two transcriptional regulators of the SOX family, SOX17 and SOX7, within the PrE and its derivatives. We noted that SOX17 was first detected in a salt-and-pepper distribution within the ICM, subsequently becoming restricted to the nascent PrE epithelium. This dynamic distribution of SOX17 resembled the localization of GATA6 and GATA4, two other PrE lineage-specific transcription factors. By contrast, SOX7 was only detected in PrE cells positioned in contact with the blastocoel, raising the possibility that these cells are molecularly distinct. Our observations support a model of sequential GATA6 > SOX17 > GATA4 > SOX7 transcription factor activation within the PrE lineage, perhaps correlating with the consecutive periods of cell lineage 'naïvete', commitment and sorting. Furthermore our data suggest that co-expression of SOX17 and SOX7 within sorted PrE cells could account for the absence of a detectable phenotype of Sox17 mutant blastocysts. However, analysis of implantation-delayed blastocysts, revealed a role for SOX17 in the maintenance of PrE epithelial integrity, with the absence of SOX17 leading to premature delamination and migration of parietal endoderm.
Copyright © 2010. Published by Elsevier Inc.
Figures





References
-
- Adamson ED, Ayers SE. The localization and synthesis of some collagen types in developing mouse embryos. Cell. 1979;16:953–965. - PubMed
-
- Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9:1147–1157. - PubMed
-
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. - PubMed
-
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2–MAPK pathway. Dev Cell. 2006;10:615–624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

