Deconstructing the molecular portraits of breast cancer
- PMID: 21147047
- PMCID: PMC5528267
- DOI: 10.1016/j.molonc.2010.11.003
Deconstructing the molecular portraits of breast cancer
Abstract
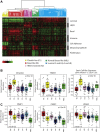
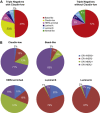
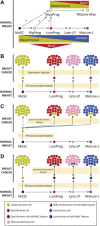
Breast cancer is a heterogeneous disease in terms of histology, therapeutic response, dissemination patterns to distant sites, and patient outcomes. Global gene expression analyses using high-throughput technologies have helped to explain much of this heterogeneity and provided important new classifications of cancer patients. In the last decade, genomic studies have established five breast cancer intrinsic subtypes (Luminal A, Luminal B, HER2-enriched, Claudin-low, Basal-like) and a Normal Breast-like group. In this review, we dissect the most recent data on this genomic classification of breast cancer with a special focus on the Claudin-low subtype, which appears enriched for mesenchymal and stem cell features. In addition, we discuss how the combination of standard clinical-pathological markers with the information provided by these genomic entities might help further understand the biological complexity of this disease, increase the efficacy of current and novel therapies, and ultimately improve outcomes for breast cancer patients.
Copyright © 2010 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures




References
-
- Agus, D.B. , Akita, R.W. , Fox, W.D. , Lewis, G.D. , Higgins, B. , Pisacane, P.I. , Lofgren, J.A. , Tindell, C. , Evans, D.P. , Maiese, K. , Scher, H.I. , Sliwkowski, M.X. , 2002. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer Cell. 2, 127 - PubMed
-
- Al Sayed, A.D. , El Weshi, A.N. , Tulbah, A.M. , Rahal, M.M. , Ezzat, A.A. , 2006. Metaplastic carcinoma of the breast clinical presentation, treatment results and prognostic factors. Acta. Oncol. 45, 188–195. - PubMed
-
- Benz, C. , Scott, G. , Sarup, J. , Johnson, R. , Tripathy, D. , Coronado, E. , Shepard, H. , Osborne, C. , 1992. Estrogen-dependent, tamoxifen-resistant tumorigenic growth of MCF-7 cells transfected with HER2/neu. Breast Cancer Res. Treat. 24, 85–95. - PubMed
-
- Carey, L.A. , Perou, C.M. , Livasy, C.A. , Dressler, L.G. , Cowan, D. , Conway, K. , Karaca, G. , Troester, M.A. , Tse, C.K. , Edmiston, S. , Deming, S.L. , Geradts, J. , Cheang, M.C. , Nielsen, T.O. , Moorman, P.G. , Earp, H.S. , Millikan, R.C. , 2006. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 295, 2492–2502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

