Notch-dependent expression of the archipelago ubiquitin ligase subunit in the Drosophila eye
- PMID: 21148181
- PMCID: PMC3005602
- DOI: 10.1242/dev.054429
Notch-dependent expression of the archipelago ubiquitin ligase subunit in the Drosophila eye
Abstract
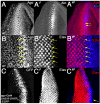
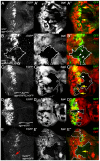
archipelago (ago)/Fbw7 encodes a conserved protein that functions as the substrate-receptor component of a polyubiquitin ligase that suppresses tissue growth in flies and tumorigenesis in vertebrates. Ago/Fbw7 targets multiple proteins for degradation, including the G1-S regulator Cyclin E and the oncoprotein dMyc/c-Myc. Despite prominent roles in growth control, little is known about the signals that regulate Ago/Fbw7 abundance in developing tissues. Here we use the Drosophila eye as a model to identify developmental signals that regulate ago expression. We find that expression of ago mRNA and protein is induced by passage of the morphogenetic furrow (MF) and identify the hedgehog (hh) and Notch (N) pathways as elements of this inductive mechanism. Cells mutant for N pathway components, or hh-defective cells that express reduced levels of the Notch ligand Delta, fail to upregulate ago transcription in the region of the MF; reciprocally, ectopic N activation in eye discs induces expression of ago mRNA. A fragment of the ago promoter that contains consensus binding sites for the N pathway transcription factor Su(H) is bound by Su(H) and confers N-inducibility in cultured cells. The failure to upregulate ago in N pathway mutant cells correlates with accumulation of the SCF-Ago target Cyclin E in the area of the MF, and this is rescued by re-expression of ago. These data suggest a model in which N acts through ago to restrict levels of the pro-mitotic factor Cyclin E. This N-Ago-Cyclin E link represents a significant new cell cycle regulatory mechanism in the developing eye.
Figures





Similar articles
-
The Drosophila ubiquitin-specific protease Puffyeye regulates dMyc-mediated growth.Development. 2013 Dec;140(23):4776-87. doi: 10.1242/dev.096941. Epub 2013 Oct 30. Development. 2013. PMID: 24173801 Free PMC article.
-
The Drosophila F box protein archipelago regulates dMyc protein levels in vivo.Curr Biol. 2004 Jun 8;14(11):965-74. doi: 10.1016/j.cub.2004.04.040. Curr Biol. 2004. PMID: 15182669
-
The archipelago ubiquitin ligase subunit acts in target tissue to restrict tracheal terminal cell branching and hypoxic-induced gene expression.PLoS Genet. 2013;9(2):e1003314. doi: 10.1371/journal.pgen.1003314. Epub 2013 Feb 14. PLoS Genet. 2013. PMID: 23459416 Free PMC article.
-
Tumor suppressor activities of the Fbw7 E3 ubiquitin ligase receptor.Cancer Biol Ther. 2005 May;4(5):506-8. doi: 10.4161/cbt.4.5.1703. Epub 2005 May 5. Cancer Biol Ther. 2005. PMID: 15908780 Review.
-
Role of the ubiquitin ligase Fbw7 in cancer progression.Cancer Metastasis Rev. 2012 Jun;31(1-2):75-87. doi: 10.1007/s10555-011-9330-z. Cancer Metastasis Rev. 2012. PMID: 22124735 Review.
Cited by
-
The tumor suppressor archipelago E3 ligase is required for spermatid differentiation in Drosophila testis.Sci Rep. 2021 Apr 19;11(1):8422. doi: 10.1038/s41598-021-87656-3. Sci Rep. 2021. PMID: 33875704 Free PMC article.
-
Requirements for mediator complex subunits distinguish three classes of notch target genes at the Drosophila wing margin.Dev Dyn. 2011 Sep;240(9):2051-9. doi: 10.1002/dvdy.22705. Epub 2011 Jul 25. Dev Dyn. 2011. PMID: 21793099 Free PMC article.
-
Novel Genes Involved in Controlling Specification of Drosophila FMRFamide Neuropeptide Cells.Genetics. 2015 Aug;200(4):1229-44. doi: 10.1534/genetics.115.178483. Epub 2015 Jun 18. Genetics. 2015. PMID: 26092715 Free PMC article.
-
Signaling pathways that control cell proliferation.Cold Spring Harb Perspect Biol. 2013 Mar 1;5(3):a008904. doi: 10.1101/cshperspect.a008904. Cold Spring Harb Perspect Biol. 2013. PMID: 23457258 Free PMC article. Review.
-
Genetic mapping of male pheromone response in the European corn borer identifies candidate genes regulating neurogenesis.Proc Natl Acad Sci U S A. 2016 Oct 18;113(42):E6401-E6408. doi: 10.1073/pnas.1610515113. Epub 2016 Oct 3. Proc Natl Acad Sci U S A. 2016. PMID: 27698145 Free PMC article.
References
-
- Bailey A. M., Posakony J. W. (1995). Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9, 2609-2622 - PubMed
-
- Baker N. E., Yu S. Y. (1998). The R8-photoreceptor equivalence group in Drosophila: fate choice precedes regulated Delta transcription and is independent of Notch gene dose. Mech. Dev. 74, 3-14 - PubMed
-
- Baker N. E., Yu S., Han D. (1996). Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr. Biol. 6, 1290-1301 - PubMed
-
- Baonza A., Freeman M. (2005). Control of cell proliferation in the Drosophila eye by Notch signaling. Dev. Cell 8, 529-539 - PubMed
-
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

