Mugsy: fast multiple alignment of closely related whole genomes
- PMID: 21148543
- PMCID: PMC3031037
- DOI: 10.1093/bioinformatics/btq665
Mugsy: fast multiple alignment of closely related whole genomes
Abstract
Motivation: The relative ease and low cost of current generation sequencing technologies has led to a dramatic increase in the number of sequenced genomes for species across the tree of life. This increasing volume of data requires tools that can quickly compare multiple whole-genome sequences, millions of base pairs in length, to aid in the study of populations, pan-genomes, and genome evolution.
Results: We present a new multiple alignment tool for whole genomes named Mugsy. Mugsy is computationally efficient and can align 31 Streptococcus pneumoniae genomes in less than 2 hours producing alignments that compare favorably to other tools. Mugsy is also the fastest program evaluated for the multiple alignment of assembled human chromosome sequences from four individuals. Mugsy does not require a reference sequence, can align mixtures of assembled draft and completed genome data, and is robust in identifying a rich complement of genetic variation including duplications, rearrangements, and large-scale gain and loss of sequence.
Availability: Mugsy is free, open-source software available from http://mugsy.sf.net.
Figures

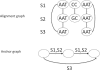
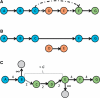

References
-
- Batzoglou S. The many faces of sequence alignment. Brief Bioinform. 2005;6:6–22. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

