The human glucocorticoid receptor as an RNA-binding protein: global analysis of glucocorticoid receptor-associated transcripts and identification of a target RNA motif
- PMID: 21148795
- PMCID: PMC3017228
- DOI: 10.4049/jimmunol.1001794
The human glucocorticoid receptor as an RNA-binding protein: global analysis of glucocorticoid receptor-associated transcripts and identification of a target RNA motif
Abstract
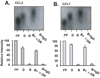
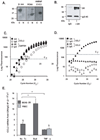
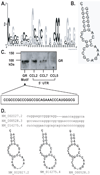
Posttranscriptional regulation is emerging as a key factor in glucocorticoid (GC)-mediated gene regulation. We investigated the role of the human GC receptor (GR) as an RNA-binding protein and its effect on mRNA turnover in human airway epithelial cells. Cell treatment with the potent GC budesonide accelerated the decay of CCL2 mRNA (t(1/2) = 8 ± 1 min versus 62 ± 17 min in DMSO-treated cells) and CCL7 mRNA (t(1/2) = 15 ± 4 min versus 114 ± 37 min), but not that of CCL5 mRNA (t(1/2)=231 ± 8 min versus 266 ± 5 min) in the BEAS-2B cell line. This effect was inhibited by preincubation with an anti-GR Ab, indicating that GR itself plays a role in the turnover of these transcripts. Coimmunoprecipitation and biotin pulldown experiments showed that GR associates with CCL2 and CCL7 mRNAs, but not CCL5 mRNA. These methods confirmed CCL2 mRNA targeting by GR in human primary airway epithelial cells. Association of the GR was localized to the 5' untranslated region of CCL2 mRNA and further mapped to nt 44-60. The collection of transcripts associated with GR, identified by immunoprecipitation of GR-mRNA complexes followed by microarray analysis, revealed 479 transcripts that associated with GR. Computational analysis of the primary sequence and secondary structures of these transcripts yielded a GC-rich motif, which was shown to bind to GR in vitro. This motif was used to predict binding of GR to an additional 7889 transcripts. These results indicate that cytoplasmic GR interacts with a subset of mRNA through specific sequences and can regulate turnover rates, suggesting a novel posttranscriptional role for GR as an RNA-binding protein.
Figures




References
-
- Stellato C. Glucocorticoid actions on airway epithelial responses in immunity: functional outcomes and molecular targets. J Allergy Clin Immunol. 2007;120:1247–1263. quiz 1264-1245. - PubMed
-
- Adcock IM, Ito K, Barnes PJ. Glucocorticoids: Effects on Gene Transcription. Proc Am Thorac Soc. 2004;1:247–254. - PubMed
-
- Bhavsar P, Hew M, Khorasani N, Torrego A, Barnes PJ, Adcock I, Chung KF. Relative corticosteroid insensitivity of alveolar macrophages in severe asthma compared with non-severe asthma. Thorax. 2008;63:784–790. - PubMed
-
- Rhen T, Cidlowski JA. Antiinflammatory Action of Glucocorticoids -- New Mechanisms for Old Drugs. N Engl J Med. 2005;353:1711–1723. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

