Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells
- PMID: 21149567
- PMCID: PMC3002033
- DOI: 10.1083/jcb.201007152
Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells
Abstract
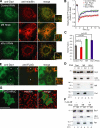
The cytoplasmic dynamin-related guanosine triphosphatase Drp1 is recruited to mitochondria and mediates mitochondrial fission. Although the mitochondrial outer membrane (MOM) protein Fis1 is thought to be a Drp1 receptor, this has not been confirmed. To analyze the mechanism of Drp1 recruitment, we manipulated the expression of mitochondrial fission and fusion proteins and demonstrated that (a) mitochondrial fission factor (Mff) knockdown released the Drp1 foci from the MOM accompanied by network extension, whereas Mff overexpression stimulated mitochondrial recruitment of Drp1 accompanied by mitochondrial fission; (b) Mff-dependent mitochondrial fission proceeded independent of Fis1; (c) a Mff mutant with the plasma membrane-targeted CAAX motif directed Drp1 to the target membrane; (d) Mff and Drp1 physically interacted in vitro and in vivo; (e) exogenous stimuli-induced mitochondrial fission and apoptosis were compromised by knockdown of Drp1 and Mff but not Fis1; and (f) conditional knockout of Fis1 in colon carcinoma cells revealed that it is dispensable for mitochondrial fission. Thus, Mff functions as an essential factor in mitochondrial recruitment of Drp1.
Figures









References
-
- Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26:211–215 10.1038/79944 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

