Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle
- PMID: 21149689
- PMCID: PMC3012510
- DOI: 10.1073/pnas.1012860108
Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle
Abstract
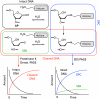
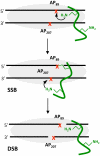
Apurinic/apyrimidinic (AP) sites are ubiquitous DNA lesions that are highly mutagenic and cytotoxic if not repaired. In addition, clusters of two or more abasic lesions within one to two turns of DNA, a hallmark of ionizing radiation, are repaired much less efficiently and thus present greater mutagenic potential. Abasic sites are chemically labile, but naked DNA containing them undergoes strand scission slowly with a half-life on the order of weeks. We find that independently generated AP sites within nucleosome core particles are highly destabilized, with strand scission occurring ∼60-fold more rapidly than in naked DNA. The majority of core particles containing single AP lesions accumulate DNA-protein cross-links, which persist following strand scission. The N-terminal region of histone protein H4 contributes significantly to DNA-protein cross-links and strand scission when AP sites are produced approximately 1.5 helical turns from the nucleosome dyad, which is a known hot spot for nucleosomal DNA damage. Reaction rates for AP sites at two positions within this region differ by ∼4-fold. However, the strand scission of the slowest reacting AP site is accelerated when it is part of a repair resistant bistranded lesion composed of two AP sites, resulting in rapid formation of double strand breaks in high yields. Multiple lysine residues within a single H4 protein catalyze double strand cleavage through a mechanism believed to involve a templating effect. These results show that AP sites within the nucleosome produce significant amounts of DNA-protein cross-links and generate double strand breaks, the most deleterious form of DNA damage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



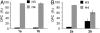

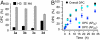
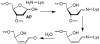
References
-
- Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: Formation, measurement and biochemical features. Mutat Res. 2003;531:5–23. - PubMed
-
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. - PubMed
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Wilson DM, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair. 2007;6:544–559. - PubMed
-
- Snowden A, Kow YW, Van Houten B. Damage repertoire of the Escherichia coli uvrabc nuclease complex includes abasic sites, base-damage analogues, and lesions containing adjacent 5′ or 3′ nicks. Biochemistry. 1990;29:7251–7259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

