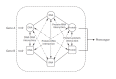
The meaning of interaction
- PMID: 21150212
- PMCID: PMC3025890
- DOI: 10.1159/000321967
The meaning of interaction
Abstract
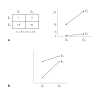
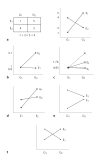
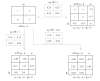
Although recent studies have attempted to dispel the confusion that exists in regard to the definition, analysis and interpretation of interaction in genetics, there still remain aspects that are poorly understood by non-statisticians. After a brief discussion of the definition of gene-gene interaction, the main part of this study addresses the fundamental meaning of statistical interaction and its relationship to measurement scale, disproportionate sample sizes in the cells of a two-way table and gametic phase disequilibrium.
Copyright © 2010 S. Karger AG, Basel.
Figures

References
-
- Musani SK, Shriner D, Liu N, Feng R, Coffey CS, Yi N, Tiwari HK, Allison DB. Detection of gene-gene interactions in genome-wide association studies of human population data. Hum Hered. 2007;63:67–84. - PubMed
-
- Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M. What is a gene, post-encode? History and updated definition. Genome Res. 2007;17:669–681. - PubMed
-
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

