The neuronal splicing factor Nova controls alternative splicing in N-type and P-type CaV2 calcium channels
- PMID: 21150296
- PMCID: PMC3047467
- DOI: 10.4161/chan.4.6.12868
The neuronal splicing factor Nova controls alternative splicing in N-type and P-type CaV2 calcium channels
Abstract
Many cellular processes are involved in optimizing protein function for specific neuronal tasks; here we focus on alternative pre-mRNA splicing. Alternative pre-mRNA splicing gives cells the capacity to modify and selectively re-balance their existing pool of transcripts in a coordinated way across multiple mRNAs, thereby effecting relatively rapid and relatively stable changes in protein activity. Here we report on and discuss the coordinated regulation of two sites of alternative splicing, e24a and e31a, in P-type CaV2.1 and N-type CaV2.2 channels. These two exons encode 4 and 2 amino acids, respectively, in the extracellular linker regions between transmembrane spanning segments S3 and S4 in domains III and IV of each CaV2 subunit. Recent genome-wide screens of splicing factor-RNA binding events by Darnell and colleagues show that Nova-2 promotes inclusion of e24a in CaV2.2 mRNAs in brain. We review these studies and show that a homologous e24a is present in theCaV2 .1 gene, Cacna1a, and that it is expressed in different regions of the nervous system. Nova-2 enhances inclusion of e24a but represses e31a inclusion in CaV2.1 and CaV2.2 mRNAs in brain. It is likely that coordinated alternative pre-mRNA splicing across related CaV2 genes by common splicing factors, allows neurons to orchestrate changes in synaptic protein function while maintaining a balanced and functioning system.
Figures
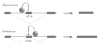

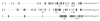


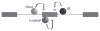
References
-
- Le Hir H, Seraphin B. EJCs at the heart of translational control. Cell. 2008;133:213–216. - PubMed
-
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. - PubMed
-
- McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
