Proteomic profiling of Myc-associated proteins
- PMID: 21150319
- PMCID: PMC3047814
- DOI: 10.4161/cc.9.24.14199
Proteomic profiling of Myc-associated proteins
Abstract
Mammalian c-Myc is a member of a small family of three closely related transcription factors. The Myc family of proto-oncogenes are among the most potent activators of tumorigenesis, and are frequently overexpressed in diverse cancers. c-Myc has an unusually broad array of regulatory functions, which include, in addition to roles in the cell cycle and apoptosis, effects on a variety of metabolic functions, cell differentiation, senescence, and stem cell maintenance. A significant number of c-Myc interacting proteins have already been defined, but it is widely believed that the c-Myc interactome is vastly larger than currently documented. In addition to interactions with components of the transcription machinery, transcription independent nuclear interactions with the DNA replication and RNA processing pathways have been reported. Cytoplasmic roles of c-Myc have also been recently substantiated. Recent advances in proteomics have opened new possibilities for the isolation of protein complexes under native conditions and confidently identifying the components using ultrasensitive, high mass accuracy and high resolution mass spectrometry techniques. In this communication we report a new tandem affinity purification (TAP) c-Myc interaction screen that employed new cell lines with near-physiological levels of c-Myc expression with multi-dimensional protein identification techniques (MudPIT) for the detection and quantification of proteins. Both label-free and the recently developed stable isotope labeling with amino acids in cell culture (SILAC) methodologies were used. Combined data from multiple biological replicates provided a dataset of 418 non-redundant proteins, 389 of which are putative novel interactors. This new information should significantly advance our understanding of this interesting and important master regulator.
Figures



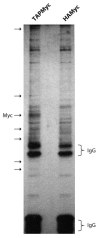

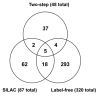
Comment in
-
More than MAX: Discovering the Myc interactome.Cell Cycle. 2011 Feb 1;10(3):374-5. doi: 10.4161/cc.10.3.14645. Epub 2011 Feb 1. Cell Cycle. 2011. PMID: 21270515 No abstract available.
References
-
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. - PubMed
-
- Albihn A, Johnsen JI, Henriksson MA. MYC in oncogenesis and as a target for cancer therapies. Adv Cancer Res. 2010;107:163–224. - PubMed
-
- Ponzielli R, Katz S, Barsyte-Lovejoy D, Penn LZ. Cancer therapeutics: Targeting the dark side of Myc. Eur J Cancer. 2005;41:2485–2501. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
