Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets
- PMID: 21151032
- PMCID: PMC7097807
- DOI: 10.1038/nrd3321
Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets
Abstract
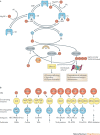
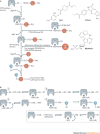
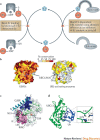
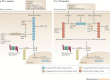
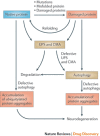
The ubiquitin-proteasome system (UPS) and ubiquitin-like protein (UBL) conjugation pathways are integral to cellular protein homeostasis. The growing recognition of the fundamental importance of these pathways to normal cell function and in disease has prompted an in-depth search for small-molecule inhibitors that selectively block the function of these pathways. However, our limited understanding of the molecular mechanisms and biological consequences of UBL conjugation is a significant hurdle to identifying drug-like inhibitors of enzyme targets within these pathways. Here, we highlight recent advances in understanding the role of some of these enzymes and how these new insights may be the key to developing novel therapeutics for diseases including immuno-inflammatory disorders, cancer, infectious diseases, cardiovascular disease and neurodegenerative disorders.
Conflict of interest statement
Lawrence R. Dick and James E. Brownell are employees of Millennium Pharmaceuticals. Lynn Bedford, James Lowe and R. John Mayer declare no competing financial interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

