Transcription of AAT•ATT triplet repeats in Escherichia coli is silenced by H-NS and IS1E transposition
- PMID: 21151567
- PMCID: PMC3000339
- DOI: 10.1371/journal.pone.0014271
Transcription of AAT•ATT triplet repeats in Escherichia coli is silenced by H-NS and IS1E transposition
Abstract
Background: The trinucleotide repeats AAT•ATT are simple DNA sequences that potentially form different types of non-B DNA secondary structures and cause genomic instabilities in vivo.
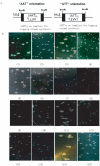
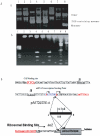
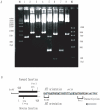
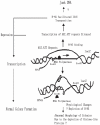
Methodology and principal findings: The molecular mechanism underlying the maintenance of a 24-triplet AAT•ATT repeat was examined in E. coli by cloning the repeats into the EcoRI site in plasmid pUC18 and into the attB site on the E. coli genome. Either the AAT or the ATT strand acted as lagging strand template in a replication fork. Propagations of the repeats in either orientation on plasmids did not affect colony morphology when triplet repeat transcription using the lacZ promoter was repressed either by supplementing LacI(Q)in trans or by adding glucose into the medium. In contrast, transparent colonies were formed by inducing transcription of the repeats, suggesting that transcription of AAT•ATT repeats was toxic to cell growth. Meanwhile, significant IS1E transposition events were observed both into the triplet repeats region proximal to the promoter side, the promoter region of the lacZ gene, and into the AAT•ATT region itself. Transposition reversed the transparent colony phenotype back into healthy, convex colonies. In contrast, transcription of an 8-triplet AAT•ATT repeat in either orientation on plasmids did not produce significant changes in cell morphology and did not promote IS1E transposition events. We further found that a role of IS1E transposition into plasmids was to inhibit transcription through the repeats, which was influenced by the presence of the H-NS protein, but not of its paralogue StpA.
Conclusions and significance: Our findings thus suggest that the longer AAT•ATT triplet repeats in E. coli become vulnerable after transcription. H-NS and its facilitated IS1E transposition can silence long triplet repeats transcription and preserve cell growth and survival.
Conflict of interest statement
Figures


Similar articles
-
Relationship between Escherichia coli growth and deletions of CTG.CAG triplet repeats in plasmids.J Mol Biol. 1996 Nov 22;264(1):82-96. doi: 10.1006/jmbi.1996.0625. J Mol Biol. 1996. PMID: 8950269
-
Interaction of DAPI with individual strands of trinucleotide repeats. Effects of replication in vitro of the AAT x ATT triplet.Eur J Biochem. 2003 Dec;270(23):4755-61. doi: 10.1046/j.1432-1033.2003.03877.x. Eur J Biochem. 2003. PMID: 14622264
-
Trinucleotide repeats affect DNA replication in vivo.Nat Genet. 1997 Nov;17(3):298-304. doi: 10.1038/ng1197-298. Nat Genet. 1997. PMID: 9354793
-
Mechanism of trinucleotide repeats instabilities: the necessities of repeat non-B secondary structure formation and the roles of cellular trans-acting factors.Yi Chuan Xue Bao. 2006 Jan;33(1):1-11. doi: 10.1016/S0379-4172(06)60001-2. Yi Chuan Xue Bao. 2006. PMID: 16450581 Review.
-
The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders.Prog Nucleic Acid Res Mol Biol. 2001;66:159-202. doi: 10.1016/s0079-6603(00)66029-4. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11051764 Review.
Cited by
-
Stress-induced modulators of repeat instability and genome evolution.J Mol Microbiol Biotechnol. 2011;21(1-2):36-44. doi: 10.1159/000332748. Epub 2012 Jan 13. J Mol Microbiol Biotechnol. 2011. PMID: 22248541 Free PMC article. Review.
-
Evaluation of mechanisms that may generate DNA lesions triggering antigenic variation in African trypanosomes.PLoS Pathog. 2018 Nov 15;14(11):e1007321. doi: 10.1371/journal.ppat.1007321. eCollection 2018 Nov. PLoS Pathog. 2018. PMID: 30440029 Free PMC article. Review.
-
Ribonuclease H1-targeted R-loops in surface antigen gene expression sites can direct trypanosome immune evasion.PLoS Genet. 2018 Dec 13;14(12):e1007729. doi: 10.1371/journal.pgen.1007729. eCollection 2018 Dec. PLoS Genet. 2018. PMID: 30543624 Free PMC article.
References
-
- Astolfi P, Bellizzi D, Sgaramella V. Frequency and coverage of trinucleotide repeats in eukaryotes. Gene. 2003;317:117–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

