Structural analysis of HIV-1 maturation using cryo-electron tomography
- PMID: 21151640
- PMCID: PMC2999899
- DOI: 10.1371/journal.ppat.1001215
Structural analysis of HIV-1 maturation using cryo-electron tomography
Abstract
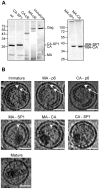
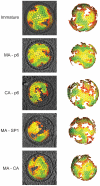
HIV-1 buds form infected cells in an immature, non-infectious form. Maturation into an infectious virion requires proteolytic cleavage of the Gag polyprotein at five positions, leading to a dramatic change in virus morphology. Immature virions contain an incomplete spherical shell where Gag is arranged with the N-terminal MA domain adjacent to the membrane, the CA domain adopting a hexameric lattice below the membrane, and beneath this, the NC domain and viral RNA forming a disordered layer. After maturation, NC and RNA are condensed within the particle surrounded by a conical CA core. Little is known about the sequence of structural changes that take place during maturation, however. Here we have used cryo-electron tomography and subtomogram averaging to resolve the structure of the Gag lattice in a panel of viruses containing point mutations abolishing cleavage at individual or multiple Gag cleavage sites. These studies describe the structural intermediates correlating with the ordered processing events that occur during the HIV-1 maturation process. After the first cleavage between SP1 and NC, the condensed NC-RNA may retain a link to the remaining Gag lattice. Initiation of disassembly of the immature Gag lattice requires cleavage to occur on both sides of CA-SP1, while assembly of the mature core also requires cleavage of SP1 from CA.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

