Complex and multidimensional lipid raft alterations in a murine model of Alzheimer's disease
- PMID: 21151659
- PMCID: PMC2997345
- DOI: 10.4061/2010/604792
Complex and multidimensional lipid raft alterations in a murine model of Alzheimer's disease
Abstract
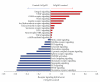
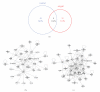
Various animal models of Alzheimer's disease (AD) have been created to assist our appreciation of AD pathophysiology, as well as aid development of novel therapeutic strategies. Despite the discovery of mutated proteins that predict the development of AD, there are likely to be many other proteins also involved in this disorder. Complex physiological processes are mediated by coherent interactions of clusters of functionally related proteins. Synaptic dysfunction is one of the hallmarks of AD. Synaptic proteins are organized into multiprotein complexes in high-density membrane structures, known as lipid rafts. These microdomains enable coherent clustering of synergistic signaling proteins. We have used mass analytical techniques and multiple bioinformatic approaches to better appreciate the intricate interactions of these multifunctional proteins in the 3xTgAD murine model of AD. Our results show that there are significant alterations in numerous receptor/cell signaling proteins in cortical lipid rafts isolated from 3xTgAD mice.
Figures






References
-
- Ashford JW. APOE genotype effects on Alzheimer’s disease onset and epidemiology. Journal of Molecular Neuroscience. 2004;23(3):157–165. - PubMed
-
- Mattson MP, Maudsley S, Martin B. BDNF and 5-HT: a dynamic duo in age-related neuronal plasticity and neurodegenerative disorders. Trends in Neurosciences. 2004;27(10):589–594. - PubMed
-
- Hardy J, Duff K, Hardy KG, Perez-Tur J, Hutton M. Genetic dissection of Alzheimer’s disease and related dementias: amyloid and its relationship to tau. Nature Neuroscience. 1998;1(5):355–358. - PubMed
-
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. - PubMed
-
- Maudsley S, Mattson MP. Protein twists and turns in Alzheimer disease. Nature Medicine. 2006;12(4):392–393. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

