Stressed-induced TMEM135 protein is part of a conserved genetic network involved in fat storage and longevity regulation in Caenorhabditis elegans
- PMID: 21151927
- PMCID: PMC2997067
- DOI: 10.1371/journal.pone.0014228
Stressed-induced TMEM135 protein is part of a conserved genetic network involved in fat storage and longevity regulation in Caenorhabditis elegans
Abstract
Disorders of mitochondrial fat metabolism lead to sudden death in infants and children. Although survival is possible, the underlying molecular mechanisms which enable this outcome have not yet been clearly identified. Here we describe a conserved genetic network linking disorders of mitochondrial fat metabolism in mice to mechanisms of fat storage and survival in Caenorhabditis elegans (C. elegans). We have previously documented a mouse model of mitochondrial very-long chain acyl-CoA dehydrogenase (VLCAD) deficiency. We originally reported that the mice survived birth, but, upon exposure to cold and fasting stresses, these mice developed cardiac dysfunction, which greatly reduced survival. We used cDNA microarrays to outline the induction of several markers of lipid metabolism in the heart at birth in surviving mice. We hypothesized that the induction of fat metabolism genes in the heart at birth is part of a regulatory feedback circuit that plays a critical role in survival. The present study uses a dual approach employing both C57BL/6 mice and the nematode, C. elegans, to focus on TMEM135, a conserved protein which we have found to be upregulated 4.3 (±0.14)-fold in VLCAD-deficient mice at birth. Our studies have demonstrated that TMEM135 is highly expressed in mitochondria and in fat-loaded tissues in the mouse. Further, when fasting and cold stresses were introduced to mice, we observed 3.25 (±0.03)- and 8.2 (±0.31)-fold increases in TMEM135 expression in the heart, respectively. Additionally, we found that deletion of the tmem135 orthologue in C. elegans caused a 41.8% (±2.8%) reduction in fat stores, a reduction in mitochondrial action potential and decreased longevity of the worm. In stark contrast, C. elegans transgenic animals overexpressing TMEM-135 exhibited increased longevity upon exposure to cold stress. Based on these results, we propose that TMEM135 integrates biological processes involving fat metabolism and energy expenditure in both the worm (invertebrates) and in mammalian organisms. The data obtained from our experiments suggest that TMEM135 is part of a regulatory circuit that plays a critical role in the survival of VLCAD-deficient mice and perhaps in other mitochondrial genetic defects of fat metabolism as well.
Conflict of interest statement
Figures
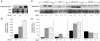



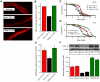

References
-
- Exil VJ, Roberts RL, Sims H, McLaughlin JE, Malkin RA, et al. Very-Long-Chain Acyl-Coenzyme A Dehydrogenase Deficiency in Mice. Circ Res. 2003;93:448–455. - PubMed
-
- Hedge P, QI R, Abernathy K, Gay C, Dharap S, et al. A Concise Guide to cDNA Microarray Analysis. Bio Techniques. 2000;29:548–562. - PubMed
-
- Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. - PubMed
-
- Bell J, Eddy B, Honkanen P, Weiner N, Woolaver T, et al. 71 A versatile system enabling analysis of slide-based high density microarrays with a variety of alternative chemistries; 1999; Nature, America, Scottsdale, AZ.
-
- Mathur A, Sims HF, Gopalakrishnan D, Gibson B, Reinaldo P, et al. Molecular Heterogeneity in Very-Long-Chain Acyl-CoA Dehydrogenase Deficiency causing Pediatric Cardiomyopathy and sudden death. Circulation. 1999;99:1337–1343. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

