Gene expression network reconstruction by convex feature selection when incorporating genetic perturbations
- PMID: 21152011
- PMCID: PMC2996324
- DOI: 10.1371/journal.pcbi.1001014
Gene expression network reconstruction by convex feature selection when incorporating genetic perturbations
Abstract
Cellular gene expression measurements contain regulatory information that can be used to discover novel network relationships. Here, we present a new algorithm for network reconstruction powered by the adaptive lasso, a theoretically and empirically well-behaved method for selecting the regulatory features of a network. Any algorithms designed for network discovery that make use of directed probabilistic graphs require perturbations, produced by either experiments or naturally occurring genetic variation, to successfully infer unique regulatory relationships from gene expression data. Our approach makes use of appropriately selected cis-expression Quantitative Trait Loci (cis-eQTL), which provide a sufficient set of independent perturbations for maximum network resolution. We compare the performance of our network reconstruction algorithm to four other approaches: the PC-algorithm, QTLnet, the QDG algorithm, and the NEO algorithm, all of which have been used to reconstruct directed networks among phenotypes leveraging QTL. We show that the adaptive lasso can outperform these algorithms for networks of ten genes and ten cis-eQTL, and is competitive with the QDG algorithm for networks with thirty genes and thirty cis-eQTL, with rich topologies and hundreds of samples. Using this novel approach, we identify unique sets of directed relationships in Saccharomyces cerevisiae when analyzing genome-wide gene expression data for an intercross between a wild strain and a lab strain. We recover novel putative network relationships between a tyrosine biosynthesis gene (TYR1), and genes involved in endocytosis (RCY1), the spindle checkpoint (BUB2), sulfonate catabolism (JLP1), and cell-cell communication (PRM7). Our algorithm provides a synthesis of feature selection methods and graphical model theory that has the potential to reveal new directed regulatory relationships from the analysis of population level genetic and gene expression data.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
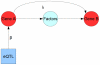
 . This gene in turn has an effect on Gene B through an unobserved pathway represented by the ‘Factors’ node. While these factors are unobserved we can still infer that there is a regulatory effect of Gene A on the downstream Gene B, which is represented in our network model by the parameter
. This gene in turn has an effect on Gene B through an unobserved pathway represented by the ‘Factors’ node. While these factors are unobserved we can still infer that there is a regulatory effect of Gene A on the downstream Gene B, which is represented in our network model by the parameter  .
.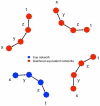





 ) using the adaptive lasso algorithm for a Saccharomyces cerevisiae cross between a wild strain and lab strain , with 112 segregants (see text for details). (a) Recovered undirected network among these 35 gene expression products and (b) putative directed network reconstructed for the same genes, based on the edges between cis-eQTL (not shown) and the 35 genes. Bold edges represent directed edges with strong confidence based on a resampling procedure (see text for details).
) using the adaptive lasso algorithm for a Saccharomyces cerevisiae cross between a wild strain and lab strain , with 112 segregants (see text for details). (a) Recovered undirected network among these 35 gene expression products and (b) putative directed network reconstructed for the same genes, based on the edges between cis-eQTL (not shown) and the 35 genes. Bold edges represent directed edges with strong confidence based on a resampling procedure (see text for details).References
-
- Emilsson V, Thorleifsson G, Zhang B, Leonardson A, Zink F, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. - PubMed
-
- Friedman N, Linial M, Nachman I, Pe'er D. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7:601–620. - PubMed
-
- Pe'er D, Regev A, Elidan G, Friedman N. Inferring subnetworks from perturbed expression profiles. Bioinformatics. 2001;17:S215. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

