Multi locus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species
- PMID: 21152037
- PMCID: PMC2996290
- DOI: 10.1371/journal.pone.0014179
Multi locus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species
Abstract
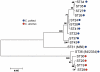
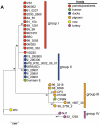
Chlamydia comprises a group of obligate intracellular bacterial parasites responsible for a variety of diseases in humans and animals, including several zoonoses. Chlamydia trachomatis causes diseases such as trachoma, urogenital infection and lymphogranuloma venereum with severe morbidity. Chlamydia pneumoniae is a common cause of community-acquired respiratory tract infections. Chlamydia psittaci, causing zoonotic pneumonia in humans, is usually hosted by birds, while Chlamydia abortus, causing abortion and fetal death in mammals, including humans, is mainly hosted by goats and sheep. We used multi-locus sequence typing to asses the population structure of Chlamydia. In total, 132 Chlamydia isolates were analyzed, including 60 C. trachomatis, 18 C. pneumoniae, 16 C. abortus, 34 C. psittaci and one of each of C. pecorum, C. caviae, C. muridarum and C. felis. Cluster analyses utilizing the Neighbour-Joining algorithm with the maximum composite likelihood model of concatenated sequences of 7 housekeeping fragments showed that C. psittaci 84/2334 isolated from a parrot grouped together with the C. abortus isolates from goats and sheep. Cluster analyses of the individual alleles showed that in all instances C. psittaci 84/2334 formed one group with C. abortus. Moving 84/2334 from the C. psittaci group to the C. abortus group resulted in a significant increase in the number of fixed differences and elimination of the number of shared mutations between C. psittaci and C. abortus. C. psittaci M56 from a muskrat branched separately from the main group of C. psittaci isolates. C. psittaci genotypes appeared to be associated with host species. The phylogenetic tree of C. psittaci did not follow that of its host bird species, suggesting host species jumps. In conclusion, we report for the first time an association between C. psittaci genotypes with host species.
Conflict of interest statement
Figures

References
-
- Everett KDE, Bush RM, Andersen AA. Emended description of the order Chlamydiales, proposal of ParaChlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49:415–440. - PubMed
-
- Stephens RS, Myers G, Eppinger M, Bavoil PM. Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol Med Microbiol. 2009;55:115–119. - PubMed
-
- White JA. Manifestations and management of lymphogranuloma venereum. Curr Opin Infect Dis. 2009;22:57–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

