Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome
- PMID: 21152079
- PMCID: PMC2994829
- DOI: 10.1371/journal.pone.0015074
Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome
Abstract
Background: Despite many attempts to establish pre-treatment prognostic markers to understand the clinical biology of esophageal adenocarcinoma (EAC), validated clinical biomarkers or parameters remain elusive. We generated and analyzed tumor transcriptome to develop a practical biomarker prognostic signature in EAC.
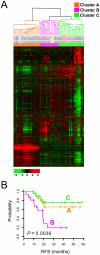
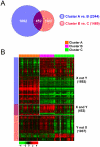
Methodology/principal findings: Untreated esophageal endoscopic biopsy specimens were obtained from 64 patients undergoing surgery and chemoradiation. Using DNA microarray technology, genome-wide gene expression profiling was performed on 75 untreated cancer specimens from 64 EAC patients. By applying various statistical and informatical methods to gene expression data, we discovered distinct subgroups of EAC with differences in overall gene expression patterns and identified potential biomarkers significantly associated with prognosis. The candidate marker genes were further explored in formalin-fixed, paraffin-embedded tissues from an independent cohort (52 patients) using quantitative RT-PCR to measure gene expression. We identified two genes whose expression was associated with overall survival in 52 EAC patients and the combined 2-gene expression signature was independently associated with poor outcome (P<0.024) in the multivariate Cox hazard regression analysis.
Conclusions/significance: Our findings suggest that the molecular gene expression signatures are associated with prognosis of EAC patients and can be assessed prior to any therapy. This signature could provide important improvement for the management of EAC patients.
Conflict of interest statement
Figures

References
-
- Bollschweiler E, Wolfgarten E, Gutschow C, Holscher AH. Demographic variations in the rising incidence of esophageal adenocarcinoma in white males. Cancer. 2001;92:549–555. - PubMed
-
- Mariette C, Balon JM, Piessen G, Fabre S, Van SI, et al. Pattern of recurrence following complete resection of esophageal carcinoma and factors predictive of recurrent disease. Cancer. 2003;97:1616–1623. - PubMed
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. - PubMed
-
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

