Viral oncogenes, noncoding RNAs, and RNA splicing in human tumor viruses
- PMID: 21152115
- PMCID: PMC2999850
- DOI: 10.7150/ijbs.6.730
Viral oncogenes, noncoding RNAs, and RNA splicing in human tumor viruses
Abstract
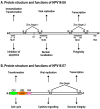
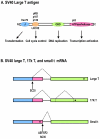
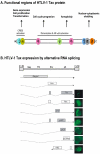
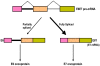
Viral oncogenes are responsible for oncogenesis resulting from persistent virus infection. Although different human tumor viruses express different viral oncogenes and induce different tumors, their oncoproteins often target similar sets of cellular tumor suppressors or signal pathways to immortalize and/or transform infected cells. Expression of the viral E6 and E7 oncogenes in papillomavirus, E1A and E1B oncogenes in adenovirus, large T and small t antigen in polyomavirus, and Tax oncogene in HTLV-1 are regulated by alternative RNA splicing. However, this regulation is only partially understood. DNA tumor viruses also encode noncoding RNAs, including viral microRNAs, that disturb normal cell functions. Among the determined viral microRNA precursors, EBV encodes 25 from two major clusters (BART and BHRF1), KSHV encodes 12 from a latent region, human polyomavirus MCV produce only one microRNA from the late region antisense to early transcripts, but HPVs appears to produce no viral microRNAs.
Keywords: Epstein-Barr virus; Human papillomaviruses; Kaposi sarcoma-associated herpesvirus; RNA splicing; adenovirus; human T-cell leukemia virus; polyomavirus; viral microRNA; viral noncoding RNA.
Conflict of interest statement
Conflict of Interests: The author has declared that no conflict of interest exists.
Figures



References
-
- Pagano JS, Blaser M, Buendia MA. et al. Infectious agents and cancer: criteria for a causal relation. Semin Cancer Biol. 2004;14:453–471. - PubMed
-
- Poiesz BJ, Ruscetti FW, Reitz MS. et al. Isolation of a new type C retrovirus (HTLV) in primary uncultured cells of a patient with Sezary T-cell leukaemia. Nature. 1981;294:268–271. - PubMed
-
- Miyoshi I, Kubonishi I, Yoshimoto S. et al. Type C virus particles in a cord T-cell line derived by co-cultivating normal human cord leukocytes and human leukaemic T cells. Nature. 1981;294:770–771. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

