Transcriptomic analysis of cell-free fetal RNA suggests a specific molecular phenotype in trisomy 18
- PMID: 21152935
- PMCID: PMC3206603
- DOI: 10.1007/s00439-010-0923-3
Transcriptomic analysis of cell-free fetal RNA suggests a specific molecular phenotype in trisomy 18
Abstract
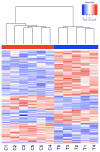
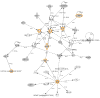
Trisomy 18 is a common human aneuploidy that is associated with significant perinatal mortality. Unlike the well-characterized "critical region" in trisomy 21 (21q22), there is no corresponding region on chromosome 18 associated with its pathogenesis. The high morbidity and mortality of affected individuals has limited extensive investigations. In order to better understand the molecular mechanisms underlying the congenital anomalies observed in this condition, we investigated the in utero gene expression profile of second trimester fetuses affected with trisomy 18. Total RNA was extracted from cell-free amniotic fluid supernatant from aneuploid fetuses and euploid controls matched for gestational age and hybridized to Affymetrix U133 Plus 2.0 arrays. Individual differentially expressed transcripts were obtained by two-tailed t tests. Over-represented functional pathways among these genes were identified with DAVID and Ingenuity(®) Pathways Analysis. Results show that three hundred and fifty-two probe sets representing 251 annotated genes were statistically significantly differentially expressed between trisomy 18 and controls. Only 7 genes (2.8% of the annotated total) were located on chromosome 18, including ROCK1, an up-regulated gene involved in valvuloseptal and endocardial cushion formation. Pathway analysis indicated disrupted function in ion transport, MHCII/T cell mediated immunity, DNA repair, G-protein mediated signaling, kinases, and glycosylation. Significant down-regulation of genes involved in adrenal development was identified, which may explain both the abnormal maternal serum estriols and the pre and postnatal growth restriction in trisomy 18. Comparison of this gene set to one previously generated for trisomy 21 fetuses revealed only six overlapping differentially regulated genes. This study contributes novel information regarding functional developmental gene expression differences in fetuses with trisomy 18.
© Springer-Verlag 2010
Figures

References
-
- Altug-Teber Ö, Walter M, Mau-Holzmann UA, Dufke A, Stappert H, Tekesin I, Heilbronner H, Nieselt K, Riess O. Specific transcriptional changes in human fetuses with autosomal trisomies. Cytogenet Genome Res. 2007;119:171–184. - PubMed
-
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, Durbin R, Falquet L, Fleischmann W, Gouzy J, Hermjakob H, Hulo N, Jonassen I, Kahn D, Kanapin A, Karavidopoulou Y, Lopez R, Marx B, Mulder NJ, Oinn TM, Pagni M, Servant F, Sigrist CJ, Zdobnov EM InterPro Consortium. InterPro: an integrated documentation resource for protein families, domains and functional sites. Bioinformatics. 2000;16:1145–1150. - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. - PMC - PubMed
-
- Baty BJ, Blackburn BL, Carey JC. Natural history of trisomy 18 and trisomy 13: I. Growth, physical assessment, medical histories, survival, and recurrence risk. Am J Med Genet. 1994;49:175–188. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

