Aligning short sequencing reads with Bowtie
- PMID: 21154709
- PMCID: PMC3010897
- DOI: 10.1002/0471250953.bi1107s32
Aligning short sequencing reads with Bowtie
Abstract
This unit shows how to use the Bowtie package to align short sequencing reads, such as those output by second-generation sequencing instruments. It also includes protocols for building a genome index and calling consensus sequences from Bowtie alignments using SAMtools.
Figures





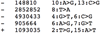
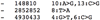
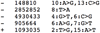
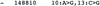
References
-
- Burrows M, Wheeler DJ. A block sorting lossless data compression algorithm. Palo Alto, CA: Digital Equipment Corporation; 1994. Technical Report 124.
-
- Ferragina P, Manzini G. Opportunistic data structures with applications; Proceedings of the 41st Annual Symposium on Foundations of Computer Science; IEEE Computer Society; 2000.
-
-
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. See above. Describes the indexing technique underlying the tool and compares to other alignment tools.
-
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

