Fabrication of stable and RNase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging
- PMID: 21155596
- PMCID: PMC3026857
- DOI: 10.1021/nn1024658
Fabrication of stable and RNase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging
Abstract
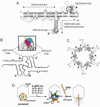
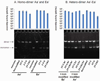
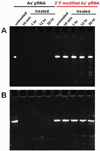
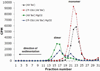
Both DNA and RNA can serve as powerful building blocks for bottom-up fabrication of nanostructures. A pioneering concept proposed by Ned Seeman 30 years ago has led to an explosion of knowledge in DNA nanotechnology. RNA can be manipulated with simplicity characteristic of DNA, while possessing noncanonical base-pairing, versatile function, and catalytic activity similar to proteins. However, standing in awe of the sensitivity of RNA to RNase degradation has made many scientists flinch away from RNA nanotechnology. Here we report the construction of stable RNA nanoparticles resistant to RNase digestion. The 2'-F (2'-fluoro) RNA retained its property for correct folding in dimer formation, appropriate structure in procapsid binding, and biological activity in gearing the phi29 nanomotor to package viral DNA and producing infectious viral particles. Our results demonstrate that it is practical to produce RNase-resistant, biologically active, and stable RNA for application in nanotechnology.
Figures






References
-
- Aldaye FA, Palmer AL, Sleiman HF. Assembling Materials With DNA As the Guide. Science. 2008;321:1795–1799. - PubMed
-
- Glotzer SC. Materials Science. Some Assembly Required. Science. 2004;306:419–420. - PubMed
-
- Gates BD, Xu Q, Stewart M, Ryan D, Willson CG, Whitesides GM. New Approaches to Nanofabrication: Molding, Printing, and Other Techniques. Chem. Rev. 2005;105:1171–1196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

