Constitutive canonical NF-κB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma
- PMID: 21156282
- PMCID: PMC3018685
- DOI: 10.1016/j.ccr.2010.11.024
Constitutive canonical NF-κB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma
Abstract
Diffuse large B cell lymphoma (DLBCL) comprises disease entities with distinct genetic profiles, including germinal center B cell (GCB)-like and activated B cell (ABC)-like DLBCLs. Major differences between these two subtypes include genetic aberrations leading to constitutive NF-κB activation and interference with terminal B cell differentiation through BLIMP1 inactivation, observed in ABC- but not GCB-DLBCL. Using conditional gain-of-function and/or loss-of-function mutagenesis in the mouse, we show that constitutive activation of the canonical NF-κB pathway cooperates with disruption of BLIMP1 in the development of a lymphoma that resembles human ABC-DLBCL. Our work suggests that both NF-κB signaling, as an oncogenic event, and BLIMP1, as a tumor suppressor, play causal roles in the pathogenesis of ABC-DLBCL.
Copyright © 2010 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

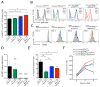



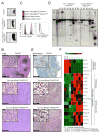
Comment in
-
BLIMP1 against lymphoma: The verdict is reached.Cancer Cell. 2010 Dec 14;18(6):537-9. doi: 10.1016/j.ccr.2010.11.029. Cancer Cell. 2010. PMID: 21156275
References
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
-
- Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J Immunol. 2000;165:5462–5471. - PubMed
-
- Banham AH, Connors JM, Brown PJ, Cordell JL, Ott G, Sreenivasan G, Farinha P, Horsman DE, Gascoyne RD. Expression of the FOXP1 transcription factor is strongly associated with inferior survival in patients with diffuse large B-cell lymphoma. Clin Cancer Res. 2005;11:1065–1072. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

