Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore
- PMID: 21157429
- PMCID: PMC3025471
- DOI: 10.1038/emboj.2010.329
Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore
Abstract
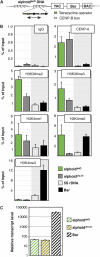
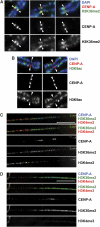
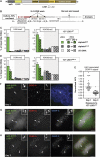
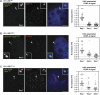
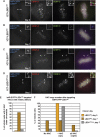
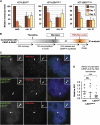
Kinetochores assemble on distinct 'centrochromatin' containing the histone H3 variant CENP-A and interspersed nucleosomes dimethylated on H3K4 (H3K4me2). Little is known about how the chromatin environment at active centromeres governs centromeric structure and function. Here, we report that centrochromatin resembles K4-K36 domains found in the body of some actively transcribed housekeeping genes. By tethering the lysine-specific demethylase 1 (LSD1), we specifically depleted H3K4me2, a modification thought to have a role in transcriptional memory, from the kinetochore of a synthetic human artificial chromosome (HAC). H3K4me2 depletion caused kinetochores to suffer a rapid loss of transcription of the underlying α-satellite DNA and to no longer efficiently recruit HJURP, the CENP-A chaperone. Kinetochores depleted of H3K4me2 remained functional in the short term, but were defective in incorporation of CENP-A, and were gradually inactivated. Our data provide a functional link between the centromeric chromatin, α-satellite transcription, maintenance of CENP-A levels and kinetochore stability.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Comment in
-
Histone H3K4 methylation keeps centromeres open for business.EMBO J. 2011 Jan 19;30(2):233-4. doi: 10.1038/emboj.2010.339. EMBO J. 2011. PMID: 21245889 Free PMC article.
References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

