Atomic model of an infectious rotavirus particle
- PMID: 21157433
- PMCID: PMC3025467
- DOI: 10.1038/emboj.2010.322
Atomic model of an infectious rotavirus particle
Abstract
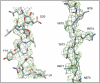
Non-enveloped viruses of different types have evolved distinct mechanisms for penetrating a cellular membrane during infection. Rotavirus penetration appears to occur by a process resembling enveloped-virus fusion: membrane distortion linked to conformational changes in a viral protein. Evidence for such a mechanism comes from crystallographic analyses of fragments of VP4, the rotavirus-penetration protein, and infectivity analyses of structure-based VP4 mutants. We describe here the structure of an infectious rotavirus particle determined by electron cryomicroscopy (cryoEM) and single-particle analysis at about 4.3 Å resolution. The cryoEM image reconstruction permits a nearly complete trace of the VP4 polypeptide chain, including the positions of most side chains. It shows how the two subfragments of VP4 (VP8(*) and VP5(*)) retain their association after proteolytic cleavage, reveals multiple structural roles for the β-barrel domain of VP5(*), and specifies interactions of VP4 with other capsid proteins. The virion model allows us to integrate structural and functional information into a coherent mechanism for rotavirus entry.
Conflict of interest statement
PRD and ECS are employees and shareholders of Novartis Vaccines and Diagnostics, Inc.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

