Insights into base selectivity from the 1.8 Å resolution structure of an RB69 DNA polymerase ternary complex
- PMID: 21158418
- PMCID: PMC3036992
- DOI: 10.1021/bi101192f
Insights into base selectivity from the 1.8 Å resolution structure of an RB69 DNA polymerase ternary complex
Abstract
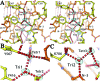
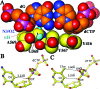
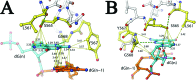
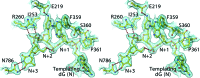
Bacteriophage RB69 DNA polymerase (RB69 pol) has served as a model for investigating how B family polymerases achieve a high level of fidelity during DNA replication. We report here the structure of an RB69 pol ternary complex at 1.8 Å resolution, extending the resolution from our previously reported structure at 2.6 Å [Franklin, M. C., et al. (2001) Cell 105, 657-667]. In the structure presented here, a network of five highly ordered, buried water molecules can be seen to interact with the N3 and O2 atoms in the minor groove of the DNA duplex. This structure reveals how the formation of the closed ternary complex eliminates two ordered water molecules, which are responsible for a kink in helix P in the apo structure. In addition, three pairs of polar-nonpolar interactions have been observed between (i) the Cα hydrogen of G568 and the N3 atom of the dG templating base, (ii) the O5' and C5 atoms of the incoming dCTP, and (iii) the OH group of S565 and the aromatic face of the dG templating base. These interactions are optimized in the dehydrated environment that envelops Watson-Crick nascent base pairs and serve to enhance base selectivity in wild-type RB69 pol.
Figures


References
-
- Echols H.; Goodman M. F. (1991) Fidelity mechanisms in DNA replication. Annu. Rev. Biochem. 60, 477–511. - PubMed
-
- Joyce C. M.; Steitz T. A. (1994) Function and structure relationship in DNA polymerases. Annu. Rev. Biochem. 63, 777–822. - PubMed
-
- Wang J.; Sattar A. K.; Wang C. C.; Karam J. D.; Konigsberg W. H.; Steitz T. A. (1997) Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell 89, 1087–1099. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

