Cohesin organizes chromatin loops at DNA replication factories
- PMID: 21159821
- PMCID: PMC3003199
- DOI: 10.1101/gad.608210
Cohesin organizes chromatin loops at DNA replication factories
Abstract
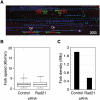
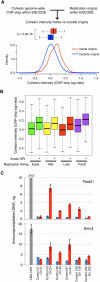
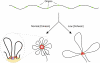
Genomic DNA is packed in chromatin fibers organized in higher-order structures within the interphase nucleus. One level of organization involves the formation of chromatin loops that may provide a favorable environment to processes such as DNA replication, transcription, and repair. However, little is known about the mechanistic basis of this structuration. Here we demonstrate that cohesin participates in the spatial organization of DNA replication factories in human cells. Cohesin is enriched at replication origins and interacts with prereplication complex proteins. Down-regulation of cohesin slows down S-phase progression by limiting the number of active origins and increasing the length of chromatin loops that correspond with replicon units. These results give a new dimension to the role of cohesin in the architectural organization of interphase chromatin, by showing its participation in DNA replication.
Figures




Comment in
-
New temptations in SMC research.Nat Rev Mol Cell Biol. 2012 Apr 4;13(5):282. doi: 10.1038/nrm3331. Nat Rev Mol Cell Biol. 2012. PMID: 22473469 No abstract available.
References
-
- Berezney R, Dubey DD, Huberman JA 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 - PubMed
-
- Borlado LR, Mendez J 2008. CDC6: From DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 29: 237–243 - PubMed
-
- Buongiorno-Nardelli M, Micheli G, Carri MT, Marilley M 1982. A relationship between replicon size and supercoiled loop domains in the eukaryotic genome. Nature 298: 100–102 - PubMed
-
- Casado-Vela J, Martinez-Torrecuadrada JL, Casal JI 2009. Differential phosphorylation patterns between the Cyclin-A2/CDK2 complex and their monomers. Protein Expr Purif 66: 15–21 - PubMed
