Inhibition of early stages of HIV-1 assembly by INI1/hSNF5 transdominant negative mutant S6
- PMID: 21159874
- PMCID: PMC3067803
- DOI: 10.1128/JVI.00006-10
Inhibition of early stages of HIV-1 assembly by INI1/hSNF5 transdominant negative mutant S6
Abstract
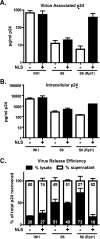
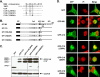
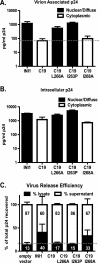
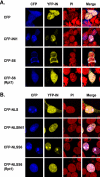
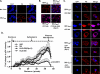
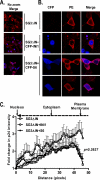
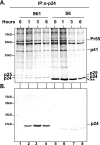
INI1/hSNF5 is an HIV-1 integrase (IN) binding protein specifically incorporated into virions. A truncated mutant of INI1 (S6, amino acids 183 to 294) harboring the minimal IN binding Rpt1 domain potently inhibits HIV-1 particle production in a transdominant manner. The inhibition requires interaction of S6 with IN within Gag-Pol. While INI1 is a nuclear protein and harbors a masked nuclear export signal (NES), the transdominant negative mutant S6 is cytoplasmic, due to the unmasking of NES. Here, we examined the effects of subcellular localization of S6 on HIV-1 inhibition and further investigated the stages of assembly that are affected. We found that targeting a nuclear localization signal-containing S6 variant [NLS-S6(Rpt1)] to the nucleoplasm (but not to the nucleolus) resulted in complete reversal of inhibition of particle production. Electron microscopy indicated that although no electron-dense particles at any stage of assembly were seen in cells expressing S6, virions were produced in cells expressing the rescue mutant NLS-S6(Rpt1) to wild-type levels. Immunofluorescence analysis revealed that p24 exhibited a diffuse pattern of localization within the cytoplasm in cells expressing S6 in contrast to accumulation along the membrane in controls. Pulse-chase analysis indicated that in S6-expressing cells, although Gag(Pr55(gag)) protein translation was unaffected, processing and release of p24 were defective. Together, these results indicate that expression of S6 in the cytoplasm interferes with trafficking of Gag-Pol/Gag to the membrane and causes a defective processing leading to inhibition of assembly at an early stage prior to particle formation and budding.
Figures


References
-
- Asp, P., M. Wihlborg, M. Karlen, and A. K. Farrants. 2002. Expression of BRG1, a human SWI/SNF component, affects the organisation of actin filaments through the RhoA signaling pathway. J. Cell Sci. 115:2735-2746. - PubMed
-
- Batonick, M., et al. 2005. Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology 342:190-200. - PubMed
-
- Brown, P. 1997. Integration, p. 161-203. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

