Serotype-specific inactivation of the cellular DNA damage response during adenovirus infection
- PMID: 21159879
- PMCID: PMC3067775
- DOI: 10.1128/JVI.01748-10
Serotype-specific inactivation of the cellular DNA damage response during adenovirus infection
Abstract
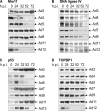
Adenovirus type 5 (Ad5) inactivates the host cell DNA damage response by facilitating the degradation of Mre11, DNA ligase IV, and p53. In the case of p53, this is achieved through polyubiquitylation by Ad5E1B55K and Ad5E4orf6, which recruit a Cul5-based E3 ubiquitin ligase. Recent evidence indicates that this paradigm does not apply to other adenovirus serotypes, since Ad12, but not Ad5, causes the degradation of TOPBP1 through the action of E4orf6 alone and a Cul2-based E3 ubiquitin ligase. We now have extended these studies to adenovirus groups A to E. While infection by Ad4, Ad5, and Ad12 (groups E, C, and A, respectively) cause the degradation of Mre11, DNA ligase IV, and p53, infection with Ad3, Ad7, Ad9, and Ad11 (groups B1, B1, D, and B2, respectively) only affects DNA ligase IV levels. Indeed, Ad3, Ad7, and Ad11 cause the marked accumulation of p53. Despite this, MDM2 levels were very low following infection with all of the viruses examined here, regardless of whether they increase p53 expression. In addition, we found that only Ad12 causes the degradation of TOPBP1, and, like Ad5, Ad4 recruits a Cul5-based E3 ubiquitin ligase to degrade p53. Surprisingly, Mre11 and DNA ligase IV degradation do not appear to be significantly affected in Ad4-, Ad5-, or Ad12-infected cells depleted of Cul2 or Cul5, indicating that E1B55K and E4orf6 recruit multiple ubiquitin ligases to target cellular proteins. Finally, although Mre11 is not degraded by Ad3, Ad7, Ad9, and Ad11, no viral DNA concatemers could be detected. We suggest that group B and D adenoviruses have evolved mechanisms based on the loss of DNA ligase IV and perhaps other unknown molecules to disable the host cell DNA damage response to promote viral replication.
Figures





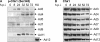


References
-
- Benko, M., B. Harrach, and W. C. Russell. 2000. Adenoviridae, p. 227-235. In M. H. Van Regenmortel, C. M. Faquert, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Malinoff, M. A. Mayo, D. J. McGeooh, C. R. Pringle, and R. B. Wickner (ed.). Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, New York, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

