Molecular determination of H antigens of Salmonella by use of a microsphere-based liquid array
- PMID: 21159932
- PMCID: PMC3043481
- DOI: 10.1128/JCM.01323-10
Molecular determination of H antigens of Salmonella by use of a microsphere-based liquid array
Abstract
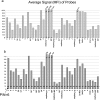
Serotyping of Salmonella has been an invaluable subtyping method for epidemiologic studies for more than 70 years. The technical difficulties of serotyping, primarily in antiserum production and quality control, can be overcome with modern molecular methods. We developed a DNA-based assay targeting the genes encoding the flagellar antigens (fliC and fljB) of the Kauffmann-White serotyping scheme. Fifteen H antigens (H:a, -b, -c, -d, -d/j, -e,h, -i, -k, -r, -y, -z, -z(10), -z(29), -z(35), and -z(6)), 5 complex major antigens (H:G, -EN, -Z4, -1, and -L) and 16 complex secondary antigens (H:2, -5, -6, -7, -f, -m/g,m, -m/m,t, -p, -s, -t/m,t, -v, -x, -z(15), -z(24), -z(28), and -z(51)) were targeted in the assay. DNA probes targeting these antigens were designed and evaluated on 500 isolates tested in parallel with traditional serotyping methods. The assay correctly identified 461 (92.2%) isolates based on the 36 antigens detected in the assay. Among the isolates considered correctly identified, 47 (9.4%) were partially serotyped because probes corresponding to some antigens in the strains were not in the assay, and 13 (2.6%) were monophasic or nonmotile strains that possessed flagellar antigen genes that were not expressed but were detected in the assay. The 39 (7.8%) strains that were not correctly identified possessed an antigen that should have been detected by the assay but was not. Apparent false-negative results may be attributed to allelic divergence. The molecular assay provided results that paralleled traditional methods with a much greater throughput, while maintaining the integrity of the Kauffmann-White serotyping scheme, thus providing backwards-compatible epidemiologic data. This assay should greatly enhance the ability of clinical and public health laboratories to serotype Salmonella.
Figures


References
-
- Department of Health and Human Services 2006. Salmonella: annual summary 2006. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/ncidod/dbmd/phlisdata/salmtab/2006/SalmonellaAnnualSu...
-
- Echeita M. A., Herrera S., Garaizar J., Usera M. A. 2002. Multiplex PCR-based detection and identification of the most common Salmonella second-phase flagellar antigens. Res. Microbiol. 153:107–113 - PubMed
-
- Ewing W. H. 1972. The nomenclature of Salmonella, its usage, and definitions for the three species. Can. J. Microbiol. 18:1629–1637 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

