Quantification of lentiviral vector copy numbers in individual hematopoietic colony-forming cells shows vector dose-dependent effects on the frequency and level of transduction
- PMID: 21160533
- PMCID: PMC3130191
- DOI: 10.1038/gt.2010.163
Quantification of lentiviral vector copy numbers in individual hematopoietic colony-forming cells shows vector dose-dependent effects on the frequency and level of transduction
Abstract
Lentiviral vectors are effective tools for gene transfer and integrate variable numbers of proviral DNA copies in variable proportions of cells. The levels of transduction of a cellular population may therefore depend upon experimental parameters affecting the frequency and/or the distribution of vector integration events in this population. Such analysis would require measuring vector copy numbers (VCN) in individual cells. To evaluate the transduction of hematopoietic progenitor cells at the single-cell level, we measured VCN in individual colony-forming cell (CFC) units, using an adapted quantitative PCR (Q-PCR) method. The feasibility, reproducibility and sensitivity of this approach were tested with characterized cell lines carrying known numbers of vector integration. The method was validated by correlating data in CFC with gene expression or with calculated values, and was found to slightly underestimate VCN. In spite of this, such Q-PCR on CFC was useful to compare transduction levels with different infection protocols and different vectors. Increasing the vector concentration and re-iterating the infection were two different strategies that improved transduction by increasing the frequency of transduced progenitor cells. Repeated infection also augmented the number of integrated copies and the magnitude of this effect seemed to depend on the vector preparation. Thus, the distribution of VCN in hematopoietic colonies may depend upon experimental conditions including features of vectors. This should be carefully evaluated in the context of ex vivo hematopoietic gene therapy studies.
Figures


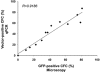
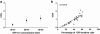
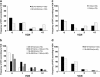
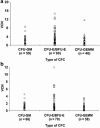
References
-
- Fischer A, Cavazzana-Calvo M. Gene therapy of inherited diseases. Lancet. 2008;371:2044–2047. - PubMed
-
- Piacibello W, Bruno S, Sanavio F, Droetto S, Gunetti M, Ailles L, et al. Lentiviral gene transfer and ex vivo expansion of human primitive stem cells capable of primary, secondary, and tertiary multilineage repopulation in NOD/SCID mice. Nonobese diabetic/severe combined immunodeficient. Blood. 2002;100:4391–4400. - PubMed
-
- Woods NB, Fahlman C, Mikkola H, Hamaguchi I, Olsson K, Zufferey R, et al. Lentiviral gene transfer into primary and secondary NOD/SCID repopulating cells. Blood. 2000;96:3725–3733. - PubMed
-
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

