Intercalation of a new tier of transcription regulation into an ancient circuit
- PMID: 21164485
- PMCID: PMC3254258
- DOI: 10.1038/nature09560
Intercalation of a new tier of transcription regulation into an ancient circuit
Abstract
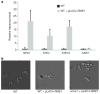
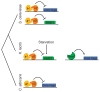
Changes in gene regulatory networks are a major source of evolutionary novelty. Here we describe a specific type of network rewiring event, one that intercalates a new level of transcriptional control into an ancient circuit. We deduce that, over evolutionary time, the direct ancestral connections between a regulator and its target genes were broken and replaced by indirect connections, preserving the overall logic of the ancestral circuit but producing a new behaviour. The example was uncovered through a series of experiments in three ascomycete yeasts: the bakers' yeast Saccharomyces cerevisiae, the dairy yeast Kluyveromyces lactis and the human pathogen Candida albicans. All three species have three cell types: two mating-competent cell forms (a and α) and the product of their mating (a/α), which is mating-incompetent. In the ancestral mating circuit, two homeodomain proteins, Mata1 and Matα2, form a heterodimer that directly represses four genes that are expressed only in a and α cells and are required for mating. In a relatively recent ancestor of K. lactis, a reorganization occurred. The Mata1-Matα2 heterodimer represses the same four genes (known as the core haploid-specific genes) but now does so indirectly through an intermediate regulatory protein, Rme1. The overall logic of the ancestral circuit is preserved (haploid-specific genes ON in a and α cells and OFF in a/α cells), but a new phenotype was produced by the rewiring: unlike S. cerevisiae and C. albicans, K. lactis integrates nutritional signals, by means of Rme1, into the decision of whether or not to mate.
Figures


Comment in
-
Evolution: How networks get new layers.Nat Rev Genet. 2011 Feb;12(2):78. doi: 10.1038/nrg2944. Epub 2010 Dec 30. Nat Rev Genet. 2011. PMID: 21191424 No abstract available.
References
-
- Carroll S. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. - PubMed
-
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. - PubMed
-
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nature Rev Genet. 2007;8:206–216. - PubMed
-
- Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

