Association of OLE RNA with bacterial membranes via an RNA-protein interaction
- PMID: 21166891
- PMCID: PMC3552494
- DOI: 10.1111/j.1365-2958.2010.07439.x
Association of OLE RNA with bacterial membranes via an RNA-protein interaction
Abstract
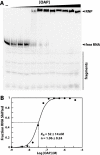
Ornate, large, extremophilic (OLE) RNAs are large, non-coding transcripts characterized by their ornate secondary structure and presence predominantly in Gram-positive, extremophilic bacteria. A gene for an OLE-associated protein (OAP) is almost always located immediately downstream of the OLE gene. OAP has no extensive homology to other proteins and is predicted to form multiple transmembrane domains. We show that this protein forms a ribonucleoprotein complex with OLE RNA using at least 2:1 protein : RNA stoichiometry. A series of truncated OLE RNA constructs was used to establish that most of the RNA can be deleted without eliminating protein binding. Two primary binding sites are present within the RNA, although additional binding determinants exist and extensive structural stabilization is induced by OAP. RNA fluorescence in situ hybridization (FISH) was used in Escherichia coli to demonstrate that ribonucleoprotein complex formation localizes the RNA near cell membranes of this heterologous system. Therefore, the majority of the complex structure formed by OLE RNA may perform a biochemical function that requires membrane localization.
© 2010 Blackwell Publishing Ltd.
Figures

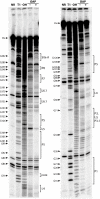
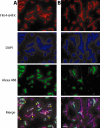
Comment in
-
Membrane RNAs in bacteria.Mol Microbiol. 2011 Jan;79(1):1-2. doi: 10.1111/j.1365-2958.2010.07438.x. Mol Microbiol. 2011. PMID: 21210530 Free PMC article. No abstract available.
References
-
- Baliarda A, Robert H, Jebbar M, Blanco C, Le Marrec C. Isolation and characterization of ButA, a secondary glycine betaine transport system operating in Tetragenococcus halophila. Curr Microbiol. 2003;47:347–351. - PubMed
-
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. - PubMed
-
- Brandt W, Brauer L, Gunnewich N, Kufka J, Rausch F, Schulze D, et al. Molecular and structural basis of metabolic diversity mediated by prenyldiphosphate converting enzymes. Phytochemistry. 2009;70:1758–1775. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

