Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence
- PMID: 21166906
- PMCID: PMC3075615
- DOI: 10.1111/j.1365-2958.2010.07445.x
Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence
Abstract
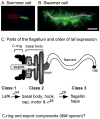
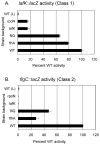
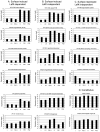
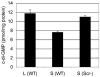
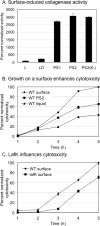
Vibrio parahaemolyticus senses surfaces via impeded rotation of its polar flagellum. We have exploited this surface-sensing mechanism to trick the organism into thinking it is on a surface when it is growing in liquid. This facilitated studies of global gene expression in a way that avoided many of the complications of surface-to-liquid comparisons, and illuminated ∼ 70 genes that respond to surface sensing per se. Almost all are surface-induced (not repressed) and encode swarming motility proteins, virulence factors or sensory enzymes involved with chemoreception and c-di-GMP signalling. Follow-up studies were performed to place the surface-responsive genes in a regulatory hierarchy. Mapping the hierarchy revealed two surprises about LafK, a transcriptional activator that until now has been considered to be the master regulator for the lateral flagellar system. First, LafK controls a more diverse set of genes than previously appreciated. Second, some laf genes are not under LafK control, which means LafK is not the master regulator after all. Additional experiments motivated by the transcriptome analyses revealed that growth on a surface lowers c-di-GMP levels and enhances cytotoxicity. Thus, we demonstrate that V. parahaemolyticus can invoke a programme of gene control upon encountering a surface and the specific identities of the surface-responsive genes are pertinent to colonization and pathogenesis.
© 2010 Blackwell Publishing Ltd.
Figures
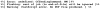

References
-
- Allison C, Emody L, Coleman N, Hughes C. The role of swarm cell differentiation and multicellular migration in the uropathogenicity of Proteus mirabilis. J Infect Dis. 1994;169:1155–1158. - PubMed
-
- An D, Parsek MR. The promise and peril of transcriptional profiling in biofilm communities. Curr Opin Microbiol. 2007;10:292–296. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Royal Statistical Soc Series B (Methodological) 1995;157:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

