XPB mediated retroviral cDNA degradation coincides with entry to the nucleus
- PMID: 21167544
- PMCID: PMC3030651
- DOI: 10.1016/j.virol.2010.11.016
XPB mediated retroviral cDNA degradation coincides with entry to the nucleus
Abstract
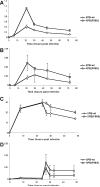
Retroviruses must integrate their cDNA to a host chromosome, but a significant fraction of retroviral cDNA is degraded before integration. XPB and XPD are part of the TFIIH complex which mediates basal transcription and DNA nucleotide excision repair. Retroviral infection increases when XPB or XPD are mutant. Here we show that inhibition of mRNA or protein synthesis does not affect HIV cDNA accumulation suggesting that TFIIH transcription activity is not required for degradation. Other host factors implicated in the stability of cDNA are not components of the XPB and XPD degradation pathway. Although an increase of retroviral cDNA in XPB or XPD mutant cells correlates with an increase of integrated provirus, the integration efficiency of pre-integration complexes is unaffected. Finally, HIV and MMLV cDNA degradation appears to coincide with nuclear import. These results suggest that TFIIH mediated cDNA degradation is a nuclear host defense against retroviral infection.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

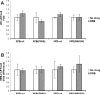
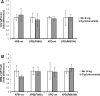

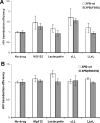
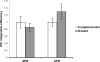
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

