Lipid-binding surfaces of membrane proteins: evidence from evolutionary and structural analysis
- PMID: 21167813
- PMCID: PMC3381425
- DOI: 10.1016/j.bbamem.2010.12.008
Lipid-binding surfaces of membrane proteins: evidence from evolutionary and structural analysis
Abstract
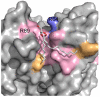
Membrane proteins function in the diverse environment of the lipid bilayer. Experimental evidence suggests that some lipid molecules bind tightly to specific sites on the membrane protein surface. These lipid molecules often act as co-factors and play important functional roles. In this study, we have assessed the evolutionary selection pressure experienced at lipid-binding sites in a set of α-helical and β-barrel membrane proteins using posterior probability analysis of the ratio of synonymous vs. nonsynonymous substitutions (ω-ratio). We have also carried out a geometric analysis of the membrane protein structures to identify residues in close contact with co-crystallized lipids. We found that residues forming cholesterol-binding sites in both β(2)-adrenergic receptor and Na(+)-K(+)-ATPase exhibit strong conservation, which can be characterized by an expanded cholesterol consensus motif for GPCRs. Our results suggest the functional importance of aromatic stacking interactions and interhelical hydrogen bonds in facilitating protein-cholesterol interactions, which is now reflected in the expanded motif. We also find that residues forming the cardiolipin-binding site in formate dehydrogenase-N γ-subunit and the phosphatidylglycerol binding site in KcsA are under strong purifying selection pressure. Although the lipopolysaccharide (LPS)-binding site in ferric hydroxamate uptake receptor (FhuA) is only weakly conserved, we show using a statistical mechanical model that LPS binds to the least stable FhuA β-strand and protects it from the bulk lipid. Our results suggest that specific lipid binding may be a general mechanism employed by β-barrel membrane proteins to stabilize weakly stable regions. Overall, we find that the residues forming specific lipid binding sites on the surfaces of membrane proteins often experience strong purifying selection pressure.
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures
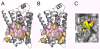

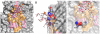

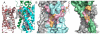

References
-
- Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. - PubMed
-
- Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. - PubMed
-
- Valiyaveetil FI, Zhou YF, Mackinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. - PubMed
-
- Cornelius F, Turner N, Christensen HRZ. Modulation of Na,K-ATPase by phospholipids and cholesterol. II. steady-state and presteady-state kinetics. Biochemistry. 2003;42:8541–8549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

