The human mitochondrial tRNAMet: structure/function relationship of a unique modification in the decoding of unconventional codons
- PMID: 21168417
- PMCID: PMC3662834
- DOI: 10.1016/j.jmb.2010.11.042
The human mitochondrial tRNAMet: structure/function relationship of a unique modification in the decoding of unconventional codons
Abstract
Human mitochondrial mRNAs utilize the universal AUG and the unconventional isoleucine AUA codons for methionine. In contrast to translation in the cytoplasm, human mitochondria use one tRNA, hmtRNA(Met)(CAU), to read AUG and AUA codons at both the peptidyl- (P-), and aminoacyl- (A-) sites of the ribosome. The hmtRNA(Met)(CAU) has a unique post-transcriptional modification, 5-formylcytidine, at the wobble position 34 (f(5)C(34)), and a cytidine substituting for the invariant uridine at position 33 of the canonical U-turn in tRNAs. The structure of the tRNA anticodon stem and loop domain (hmtASL(Met)(CAU)), determined by NMR restrained molecular modeling, revealed how the f(5)C(34) modification facilitates the decoding of AUA at the P- and the A-sites. The f(5)C(34) defined a reduced conformational space for the nucleoside, in what appears to have restricted the conformational dynamics of the anticodon bases of the modified hmtASL(Met)(CAU). The hmtASL(Met)(CAU) exhibited a C-turn conformation that has some characteristics of the U-turn motif. Codon binding studies with both Escherichia coli and bovine mitochondrial ribosomes revealed that the f(5)C(34) facilitates AUA binding in the A-site and suggested that the modification favorably alters the ASL binding kinetics. Mitochondrial translation by many organisms, including humans, sometimes initiates with the universal isoleucine codons AUU and AUC. The f(5)C(34) enabled P-site codon binding to these normally isoleucine codons. Thus, the physicochemical properties of this one modification, f(5)C(34), expand codon recognition from the traditional AUG to the non-traditional, synonymous codons AUU and AUC as well as AUA, in the reassignment of universal codons in the mitochondria.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures


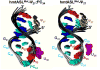

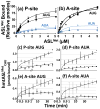
 ) and with hmtASLMetCAU-Ψ27 (
) and with hmtASLMetCAU-Ψ27 ( ). The maximum hmtASLMetCUA bound was between 3-4 pmoles. (b) AUG and AUA codon binding properties of the hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 in the A-site of the ribosome. The E. coli 70S ribosome (5 pmoles), programmed with AUG in the A-site, was titrated with hmtASLMetCAU-Ψ27;f5C34 (▲) and with hmtASLMetCAU-Ψ27 (■), or programmed with AUA and titrated with hmtASLMetCAU-Ψ27;f5C34 (
). The maximum hmtASLMetCUA bound was between 3-4 pmoles. (b) AUG and AUA codon binding properties of the hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 in the A-site of the ribosome. The E. coli 70S ribosome (5 pmoles), programmed with AUG in the A-site, was titrated with hmtASLMetCAU-Ψ27;f5C34 (▲) and with hmtASLMetCAU-Ψ27 (■), or programmed with AUA and titrated with hmtASLMetCAU-Ψ27;f5C34 ( ) and with hmtASLMetCAU-Ψ27 (
) and with hmtASLMetCAU-Ψ27 ( ). The maximum hmtASLMetCUA bound was between 0.5-1.0 pmoles. In contrast to equilibrium binding studies, the time-dependent binding of the hmtASLMetCAU-Ψ27;f5C34 (▲) and hmtASLMetCAU-Ψ27 (■) to AUG and AUA were conducted at the single concentration of ASL (0.5 μM), far too low to saturate the ribosomes. The hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 were bound to (c) AUG in the P-site of E. coli 70S ribosome, and to (d) AUA in the P-site). The hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 were bound to (e) AUG in the A-site of E. coli 70S ribosome, and to (f) AUA in the A-site.
). The maximum hmtASLMetCUA bound was between 0.5-1.0 pmoles. In contrast to equilibrium binding studies, the time-dependent binding of the hmtASLMetCAU-Ψ27;f5C34 (▲) and hmtASLMetCAU-Ψ27 (■) to AUG and AUA were conducted at the single concentration of ASL (0.5 μM), far too low to saturate the ribosomes. The hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 were bound to (c) AUG in the P-site of E. coli 70S ribosome, and to (d) AUA in the P-site). The hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 were bound to (e) AUG in the A-site of E. coli 70S ribosome, and to (f) AUA in the A-site.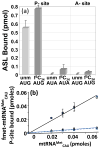
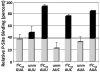
 ). Also, the AUA-programmed ribosome was titrated with either mtRNAMetCAU (◊) or its transcript (
). Also, the AUA-programmed ribosome was titrated with either mtRNAMetCAU (◊) or its transcript ( ).
).

References
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Santos MA, Moura G, Massey SE, Tuite MF. Driving change: the evolution of alternative genetic codes. Trends Genet. 2004;20:95–102. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

