Completely phased genome sequencing through chromosome sorting
- PMID: 21169219
- PMCID: PMC3017199
- DOI: 10.1073/pnas.1016725108
Completely phased genome sequencing through chromosome sorting
Erratum in
- Proc Natl Acad Sci U S A. 2012 Feb 21;109(8):3190
Abstract
The two haploid genome sequences that a person inherits from the two parents represent the most fundamentally useful type of genetic information for the study of heritable diseases and the development of personalized medicine. Because of the difficulty in obtaining long-range phase information, current sequencing methods are unable to provide this information. Here, we introduce and show feasibility of a scalable approach capable of generating genomic sequences completely phased across the entire chromosome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

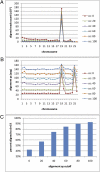
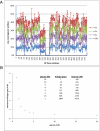
References
-
- Venter JC. Multiple personal genomes await. Nature. 2010;464:676–677. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

