Preferential repair of oxidized base damage in the transcribed genes of mammalian cells
- PMID: 21169365
- PMCID: PMC3057786
- DOI: 10.1074/jbc.M110.198796
Preferential repair of oxidized base damage in the transcribed genes of mammalian cells
Abstract
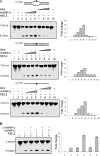
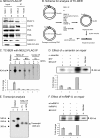
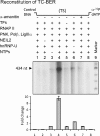
Preferential repair of bulky DNA adducts from the transcribed genes via nucleotide excision repair is well characterized in mammalian cells. However, definitive evidence is lacking for similar repair of oxidized bases, the major endogenous DNA lesions. Here we show that the oxidized base-specific human DNA glycosylase NEIL2 associates with RNA polymerase II and the transcriptional regulator heterogeneous nuclear ribonucleoprotein-U (hnRNP-U), both in vitro and in cells. NEIL2 immunocomplexes from cell extracts preferentially repaired the mutagenic cytosine oxidation product 5-hydroxyuracil in the transcribed strand. In a reconstituted system, we also observed NEIL2-initiated transcription-dependent base excision repair of 5-hydroxyuracil in the transcribed strand, with hnRNP-U playing a critical role. Chromatin immunoprecipitation/reimmunoprecipitation studies showed association of NEIL2, RNA polymerase II, and hnRNP-U on transcribed but not on transcriptionally silent genes. Furthermore, NEIL2-depleted cells accumulated more DNA damage in active than in silent genes. These results strongly support the preferential role of NEIL2 in repairing oxidized bases in the transcribed genes of mammalian cells.
Figures



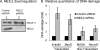
References
-
- Breimer L. H. (1990) Mol. Carcinog. 3, 188–197 - PubMed
-
- Lavrovsky Y., Chatterjee B., Clark R. A., Roy A. K. (2000) Exp. Gerontol. 35, 521–532 - PubMed
-
- Krokan H. E., Nilsen H., Skorpen F., Otterlei M., Slupphaug G. (2000) FEBS Lett. 476, 73–77 - PubMed
-
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., Ellenberger T. (2006) DNA Repair and Mutagenesis, American Society for Microbiology Press, Washington, D. C.
-
- Ikeda S., Biswas T., Roy R., Izumi T., Boldogh I., Kurosky A., Sarker A. H., Seki S., Mitra S. (1998) J. Biol. Chem. 273, 21585–21593 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

