Identifying cancer driver genes in tumor genome sequencing studies
- PMID: 21169372
- PMCID: PMC3018819
- DOI: 10.1093/bioinformatics/btq630
Identifying cancer driver genes in tumor genome sequencing studies
Abstract
Motivation: Major tumor sequencing projects have been conducted in the past few years to identify genes that contain 'driver' somatic mutations in tumor samples. These genes have been defined as those for which the non-silent mutation rate is significantly greater than a background mutation rate estimated from silent mutations. Several methods have been used for estimating the background mutation rate.
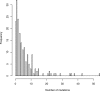
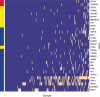
Results: We propose a new method for identifying cancer driver genes, which we believe provides improved accuracy. The new method accounts for the functional impact of mutations on proteins, variation in background mutation rate among tumors and the redundancy of the genetic code. We reanalyzed sequence data for 623 candidate genes in 188 non-small cell lung tumors using the new method. We found several important genes like PTEN, which were not deemed significant by the previous method. At the same time, we determined that some genes previously reported as drivers were not significant by the new analysis because mutations in these genes occurred mainly in tumors with large background mutation rates.
Availability: The software is available at: http://linus.nci.nih.gov/Data/YounA/software.zip.
Figures
References
-
- Agathanggelou A, et al. Epigenetic inactivation of the candidate 3p21.3 suppressor gene blu in human cancers. Oncogene. 2003;22:1580–1588. - PubMed
-
- Casella G. An introduction to empirical Bayes data analysis. Am. Stat. 1985;39:83–87.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

