Using a stem cell-based signature to guide therapeutic selection in cancer
- PMID: 21169407
- PMCID: PMC3049992
- DOI: 10.1158/0008-5472.CAN-10-1735
Using a stem cell-based signature to guide therapeutic selection in cancer
Abstract
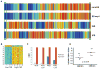
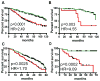
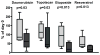
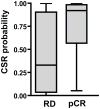
Given the very substantial heterogeneity of most human cancers, it is likely that most cancer therapeutics will be active in only a small fraction of any population of patients. As such, the development of new therapeutics, coupled with methods to match a therapy with the individual patient, will be critical to achieving significant gains in disease outcome. One such opportunity is the use of expression signatures to identify key oncogenic phenotypes that can serve not only as biomarkers but also as a means of identifying therapeutic compounds that might specifically target these phenotypes. Given the potential importance of targeting tumors exhibiting a stem-like phenotype, we have developed an expression signature that reflects common biological aspects of various stem-like characteristics. The consensus stemness ranking (CSR) signature is upregulated in cancer stem cell-enriched samples at advanced tumor stages and is associated with poor prognosis in multiple cancer types. Using two independent computational approaches we utilized the CSR signature to identify clinically useful compounds that could target the CSR phenotype. In vitro assays confirmed selectivity of several predicted compounds including topoisomerase inhibitors and resveratrol towards breast cancer cell lines that exhibit a high-CSR phenotype. Importantly, the CSR signature could predict clinical response of breast cancer patients to a neoadjuvant regimen that included a CSR-specific agent. Collectively, these results suggest therapeutic opportunities to target the CSR phenotype in a relevant cohort of cancer patients.
©2010 AACR.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

