Systematic screen of Schizosaccharomyces pombe deletion collection uncovers parallel evolution of the phosphate signal transduction pathway in yeasts
- PMID: 21169418
- PMCID: PMC3067400
- DOI: 10.1128/EC.00216-10
Systematic screen of Schizosaccharomyces pombe deletion collection uncovers parallel evolution of the phosphate signal transduction pathway in yeasts
Abstract
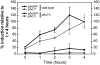
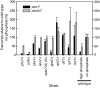
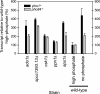
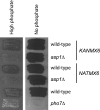
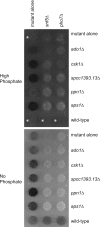
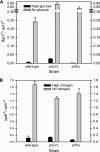
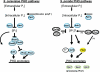
The phosphate signal transduction (PHO) pathway, which regulates genes in response to phosphate starvation, is well defined in Saccharomyces cerevisiae. We asked whether the PHO pathway was the same in the distantly related fission yeast Schizosaccharomyces pombe. We screened a deletion collection for mutants aberrant in phosphatase activity, which is primarily a consequence of pho1(+) transcription. We identified a novel zinc finger-containing protein (encoded by spbc27b12.11c(+)), which we have named pho7(+), that is essential for pho1(+) transcriptional induction during phosphate starvation. Few of the S. cerevisiae genes involved in the PHO pathway appear to be involved in the regulation of the phosphate starvation response in S. pombe. Only the most upstream genes in the PHO pathway in S. cerevisiae (ADO1, DDP1, and PPN1) share a similar role in both yeasts. Because ADO1 and DDP1 regulate ATP and IP(7) levels, we hypothesize that the ancestor of these yeasts must have sensed similar metabolites in response to phosphate starvation but have evolved distinct mechanisms in parallel to sense these metabolites and induce phosphate starvation genes.
Figures


References
-
- Barbet N., Muriel W. J., Carr A. M. 1992. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene 114:59–66 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

