The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation
- PMID: 21169563
- PMCID: PMC3098594
- DOI: 10.1074/mcp.M110.006460
The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation
Abstract
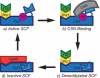
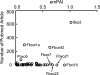
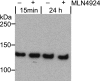
The human genome encodes 69 different F-box proteins (FBPs), each of which can potentially assemble with Skp1-Cul1-RING to serve as the substrate specificity subunit of an SCF ubiquitin ligase complex. SCF activity is switched on by conjugation of the ubiquitin-like protein Nedd8 to Cul1. Cycles of Nedd8 conjugation and deconjugation acting in conjunction with the Cul1-sequestering factor Cand1 are thought to control dynamic cycles of SCF assembly and disassembly, which would enable a dynamic equilibrium between the Cul1-RING catalytic core of SCF and the cellular repertoire of FBPs. To test this hypothesis, we determined the cellular composition of SCF complexes and evaluated the impact of Nedd8 conjugation on this steady-state. At least 42 FBPs assembled with Cul1 in HEK 293 cells, and the levels of Cul1-bound FBPs varied by over two orders of magnitude. Unexpectedly, quantitative mass spectrometry revealed that blockade of Nedd8 conjugation led to a modest increase, rather than a decrease, in the overall level of most SCF complexes. We suggest that multiple mechanisms including FBP dissociation and turnover cooperate to maintain the cellular pool of SCF ubiquitin ligases.
Figures

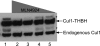
References
-
- Pickart C. M. (2001) Mechanisms underlying ubiquitination. Ann. Rev. Biochem. 70, 503–533 - PubMed
-
- Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 - PubMed
-
- Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 - PubMed
-
- Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

